Abstract
The RNase H2 complex is a conserved heterotrimeric enzyme that degrades RNA:DNA hybrids and promotes excision of rNMPs misincorporated during DNA replication. Failure to remove ribonucleotides from DNA leads to genomic instability in yeast and humans. The monogenic Aicardi-Goutières syndrome (AGS) results from mutation in one of several genes, among which are those encoding the RNase H2 subunits. The complete cellular and genomic consequences of RNASEH2 mutations and the precise connection to disease remain unclear. To learn more about the effect of RNASEH2 mutations on the cell, we used yeast as a model of AGS disease. We have generated yeast strains bearing AGS-associated mutations in RNASEH2 genes. There is a range of disease presentation in patients bearing these RNASEH2 variants. Here we report on in vivo phenotypes of genomic instability, including mutation and recombination rates, and synthetic gene interactions. These phenotypes provide insight into molecular consequences of RNASEH2 mutations, and lay the groundwork for further study of genomic instability as a contributing factor to AGS disease.
Keywords: Ribonucleotides, AGS, genome instability, recombination, mutagenesis, DNA replication
1. Introduction
Aicardi-Goutières syndrome is a recessive encephalopathy that affects the brain and immune system [1, 2]. Although affected children are born with features suggestive of a chronic prenatal viral infection [3], instead they carry mutations in one of six genes that result in an undetermined defect in processing nucleic acids and an immune response against some nucleic acid product.
1.1. RNase H2 mutations in Aicardi-Goutières syndrome patients
RNASE H2 is a conserved enzyme complex that functions in DNA replication to eliminate rNMP residues that have been incorporated into DNA and additionally can remove longer RNA:DNA hybrids or R-loops [4]. Misincorporated ribonucleotides arise either through defects in Okazaki fragment processing or as a natural consequence of errors of replicative DNA polymerases [5, 6]. Misincorporated rNMPs that are not removed distort the DNA helix and cause replication stalling in the next replication cycle. Longer RNA:DNA hybrids may arise from incomplete processing of Okazaki fragment primers or from stalled transcription complexes [4, 7].
An extensive catalog of mutations in genes leading to AGS has shown that over 50% of the studied AGS families had mutations in the RNASEH2A, RNASEH2B or RNASEH2C genes [2, 8]. Of these, two-thirds occurred in the RNASEH2B gene. Most families carried biallelic mutations, often as compound heterozygotes, with the majority of the RNASE H2 mutations occurring in the RNASEH2B gene.
1.2. DNA damage events in RNase H2-defective yeast cells
In contrast to mouse systems [5, 9], the RNASE H2 genes of yeast, RNH201, RNH202, and RNH203, are not essential, making it possible to study the DNA damage consequences of embedded ribonucleotide residues in DNA. RNase H2 null allele mutants of yeast have phenotypes associated with DNA damage and genome instability, the most common being increased mutagenesis, increased recombination, increased loss of heterozygosity (LOH), increased chromosome loss and chromosome rearrangements.
Mutational events increased in RNase H2 null mutants of yeast have a particular signature, that of slippage in simple repeats [6, 10]. These mutations are seen only in the absence of functional RNase H2 activity and require the action of Topoisomerase I (Top1) [10, 11]. Yeast RNase H2 mutants also display a hyperrecombination phenotype [12]. The hyper-recombination phenotype was not dependent on the action of Top1 and was stimulated by tandem rNMP residues in DNA, in contrast to single rNMP residues being responsible for slippage mutagenesis [13, 14]. RNase H2 mutants isolated in a different study were also found to have a hyper-recombination phenotype [15].
1.3. Studies of human AGS mutations in human and mouse cells and biochemical activity of the RNASE H2 enzyme
Identification of mutations in the RNASEH2A, RNASEH2B and RNASEH2C genes among 127 families presenting with AGS revealed 73 pedigrees with mutations [2, 16], Among the mutations observed was a homozygous mutation in RNASEH2A (c.109G>A, p.Gly37Ser) (“c” refers to the coding sequence or base which is altered and “p” refers to the amino acid residue which is altered, according to the standard nomenclature for human sequence variants [17]), predicted to affect catalytic activity, and a common mutation in RNASEH2B (c.529G>A, p.Ala177Thr), which occurred as homozygous and compound heterozygous in patients [2], Enzymatic studies confirmed that the RNASEH2A-G37S protein had reduced activity [16], Modeling this mutation into the Saccharomyces cerevisiae Rnh201 protein (Rnh201-G42S) confirmed conservation of this mutation on biochemical activity [18], Modeling other AGS alleles on the Saccharomyces cerevisiae protein were unsuccessful in that no effect on enzymatic activity in vitro could be obtained [18, 19], Of note was the rnh202-L52R allele and associated protein, which we have studied through genetic means and is described below.
A more complete understanding of the effect of AGS RNASE H2 mutations on protein function became apparent after the structure of the human RNase H2 complex was solved [20]. Mapping of 29 human AGS mutations onto the crystal structure of the complex led to predictions of effects of these mutations on complex stability and substrate cleavage. Importantly, many AGS mutations mapped to the interface between the RNase H2 subunits and would affect stability of the heterotrimer but cleavage of a rNMP containing substrate in vitro would not be affected [21], accounting for the apparent normal activity in vitro of mutant proteins. The yeast system provides the opportunity to examine the DNA damage consequences of AGS hypomorphic alleles and thereby provide support for the link between DNA damage and autoimmunity arising from nonnull RNase H2 gene mutations.
2. Materials and Methods
2.1. Yeast growth conditions
All experiments were performed by growing yeast at 30°C in either rich medium (YPD) or synthetic media plus/minus amino acids or drugs.
2.2. Plasmid and yeast strain construction
All yeast strains are derivatives of W303. Strains are listed in Supplementary Table 1. To construct AGS yeast mutants, a portion of RNH201 (−499-688) was PCR-amplified and cloned into YIplac211 integration plasmid using PstI and BamHI sites. A sequence encompassing the RNH202 locus [−388-(+)786] was PCR-amplified and cloned into YIplac211 integration plasmid using Sacl and PstI sites. Site-directed mutagenesis (QuikChange, Stratagene) was performed using the following oligos: rnh201-G42S: 5’-TCGATGAAGCTAGCAGAGGGCCCGT-3’ and 5’-ACGGGCCCTCTGCTAGCTTCATCGA-3’; rnh202-L52R: 5’-G AAAATATTAATGGAAAACGTTACGAAATAAGATC-3’ and 5’-G ATCTTATTTCGTAACGTTTTCCATTAATATTTT-3’; rnh202-H109R: 5’- CTCAAGCAAAATACGCTTTTGTTCTTTATACG −3’ and 5’-CGTATAAAGAACAAAAGCGTATTTTGCTTGAG-3’; rnh202-L186F:5’-GTCGTCTCAAGAGTGGCTTTGCAAAAGTTAGTG-3’ and 5’-CACTAACTTTTGCAAAGCCACTCTTGAGACGAC-3’; rnh202-T204l: 5’- CGTCTATTACAAGATCATATCTGCAATGATAACAC-3’ and 5’-GTGTTATCATTGCAGATATGATCTTGTAATAGACG-5’.
Mutations were confirmed by sequencing. Plasmids were linearized to transform into yeast and integrants were selected on Sc-uracil. Transformants were patched to YPD and grown overnight in YPD prior to plating on 5’FOA. FOA-resistant segregants were screened for presence of the mutation by sequencing the entire gene with some upstream and downstream regions included.
2.3. Recombination rate assay
Recombination rates were performed using the leu2-ecoRI::URA3::leu2-bstEII system as described [12], At least 18 independent cultures with a minimum of two isolates per genotype were used to determine rates and 95% confidence limits [22].
2.4. Mutation rate assay
Mutation rates for the (AG)4 slippage reporter and were determined by the Lea and Coulson median method [23] as previously described [10,11, 22].
2.5. Alkaline gel electrophoresis and quantification of ribonucleotide density
Alkaline gel analysis was performed as described [6], Genomic DNA was isolated from yeast cells grown overnight in YPD using the MasterPure Yeast DNA isolation kit (Epicentre) according to the manufacturer’s instructions. For alkaline treatment, ~10 μg purified DNA was incubated in 0.3M KOH for 2 hours at 55°C. Samples were mixed with 6X alkaline loading buffer (300 mM KOH, 6 mM EDTA, 18% Ficoll type 400, 0.15% bromocresol green, 0.25% xylene cyanol) and loaded onto a 1% agarose gel made with alkaline buffer (50 mM NaOH, 1 mM EDTA). Gels were run in 1X alkaline buffer (5 mM NaOH, 1 mM EDTA) for 18 h at 1 V/cm. Gels were neutralized by gently shaking in neutralizing buffer (1 M Tris-HCI, 1.5 M NaCI, pH 7.5) for 1 h and stained with 0.5 μg/mL ethidium bromide for 30 min, followed by destaining for 15 min in dH20. For neutral gels, ~ 2 μg purified DNA was run on 1% agarose gel 1 X TBE. Staining procedures were the same as for the alkaline gels, minus the neutralization step.
Extraction and quantification of gel data intensity profiles was carried out using Matlab code. The intensity profile (as a function of gel length) for each lane was calculated by averaging over the width of the lane followed by background subtraction. To obtain the background profile subtraction, the background profile was obtained by averaging the gap spacing on both sides of each lane. The Y-axis scale (in kb) was determined by extrapolating the specific peaks location of the different DNA fragments obtained in the intensity profile of the DNA ladder band, and this kb scale transformed onto the Y-axis of the lane profiles. The maximum of each peak was normalized to 1 and the X-axis scale are arbitrary labels to 1.
3. Results
3.1. Identification of conserved AGS-associated residues in yeast RNase H2 subunits
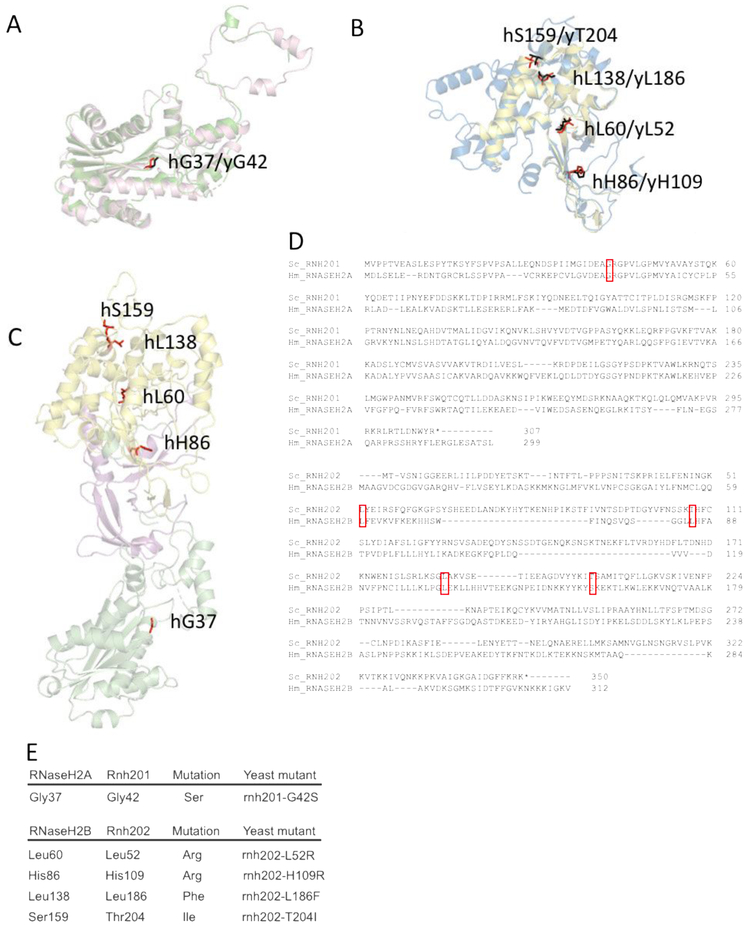
To create structural models of yeast RNaseH2 proteins, translated sequences of RNH202 and RNH201 obtained from the Saccharomyces Genome Database were submitted to the protein structure prediction server I-TASSER [24]. The resultant homology models were aligned to human RNaseH2 protein structures (PDB ID = 3PUF [20] using the COFACTOR server [25]. Images of aligned models were generated using PyMOL (Figure 1A, B). The aligned structural models were used to identify yeast residues equivalent to human residues mutated in AGS patients [2, 16]. From human RNaseH2A, residue Gly37 aligned to yeast Rnh201 residue Gly42 (Figure 1A). From human RNaseH2B, the following residues aligned to yeast Rnh202: hLeu60 to yLeu52; hHis86 to yH109; hLeu138 to yLeu186; and hSer159 to yThr204 (Figure 1B). These residues were found in various parts of the RNase H2 complex (Figure 1C). The alignments were also seen using Clustal Omega (Figure 1D). RNaseH2A is the catalytic subunit of the enzyme, and the exact functions of RNaseH2B and RNaseH2C still remain unclear, although these subunits are needed for the catalytic activity of RNASEH2A to be functional and are suggested to provide structural support as well as protein interaction platform [19]. We constructed yeast mutants bearing the same amino acid change that was seen clinically (Figure 1E), generating 5 AGS mutant strains: rnh201-G42S, rnh202-L52R, rnh202-H109R, mh202-L186F and rnh202-T204I, an assayed these AGS strains for genome instability.
Figure 1. Conserved AGS-associated residues from yeast to humans.
Homology models of yeast Rnh201 and Rnh202 were generated and aligned to structural models of human RNASEH2A and RNASEH2B, respectively. Conserved AGS-associated residues were identified on the aligned yeast homology structures. A. We chose one conserved residue in yRnh201 (magenta) / hRNASEH2A (green): yGly42(red) /hGly37(black). B. We chose four conserved residues in yRnh202(blue) / hRNASEH2B(yellow): 1.) yHis109/hHis86; 2.) yLeu52/hLeu60; 3.) yLeu186/hLeu138; 4.) yThr204/hSer159 (yeast residues in red, human residues in black). C. Conserved residues identified are located in different parts of the enzyme complex. The structure (PDB ID = 3PUFR) of the RNase H2 heterotrimeric complex consisting of RNASEH2A (green), RNASEH2B (yellow) and RNASEH2C (magenta) is shown with AGS-associated residues (red) that are conserved in yeast highlighted. D. Alignment of yeast Rnh201 with human RNASEH2A and yeast Rnh202 with human RNASEH2B. The top panel shows the alignment of Rnh201 with RNASEH2A using Clustal Omega. The lower panel shows the alignment of Rnh202 with RNASEH2B using Clustal Omega. The red boxes indicate the position of the human AGS mutations shown in panels A, B and E and their alignment with the cognate yeast residue. E. Mutations of the RNase H2 complex found in AGS patients are reconstructed in yeast strains. Yeast cells with the appropriate amino acid change that reflects AGS mutations were constructed.
3.2. Yeast AGS allele gene interactions
Deletions of any of the three RNaseH2 genes are synthetically sick/lethal with a known list of gene deletions that compromise genes involved in replication and DNA damage repair, including the genes of the MRX complex (MRE11, RAD50 and XRS2) [11], We crossed the AGS mutant strains to a rad50Δ strain, sporulated and dissected tetrads.
Two mutants, rnh201-G42S and rnh202-L52R, showed synthetic sickness with rad50A to a degree similar to that of a rnh202Δ null mutant (Figure 2A. The other three mutants, rnh202-H109R, rnh202-L186F and rnh202-T204I, did not display any synthetic sickness with the rad50Δ strain (Figure 2A. This recapitulates the in vitro data for the rnh201-G42S protein, which was shown to have reduced activity [18], However, the result with the rnh202-L52R mutant was opposite from the in vitro data, as it had been shown to have wild type activity [18].
Figure 2. Synthetic genetic interactions.
A. Synthetic gene interactions with rad50Δ. The yeast AGS mutants were crossed to a rad50Δ strain, sporulated and the resultant tetrads were dissecting in an ordered array. Genotypes of the spores were determined. Double rad50Δ rnaseH2-AGS mutants are circled in white. B. Synthetic gene interactions with rnh1Δ top1Δ. The yeast AGS mutants were crossed to a rnh1Δ top1Δ strain, sporulated and the resultant tetrads were dissecting in an ordered array. Genotypes of the spores were determined. Triple rnh1Δ top1Δ rnaseH2-AGS mutants are indicated in white squares.
We examined the interaction with deletions of both RNH1 (encoding RNase H1) and TOP1 (encoding Topisomerase I) as the triple mutant rnh1 rnh2 topi is lethal [26], In contrast to the growth defect seen with a rad50Δ mutation, only null alleles rnh20lΔ and rnh202Δ plus rnh202-L52R had a synthetic growth defect (Figure 2B). This indicates that the RNase H2 activity remaining in rnh201-G42S is sufficient to buffer against loss of RNase H1 and Top1 activities, as initially described [13]. It suggests that activity of the RNase H2 complex with the rnh202L52R subunit is below that of the complex with the rnh201G42S subunit. The structure of the human RNase H2 complex predicts that the rnaseh2B-L60R subunit impairs stability of the complex [20],
3.3. Yeast AGS alleles and genome instability assays
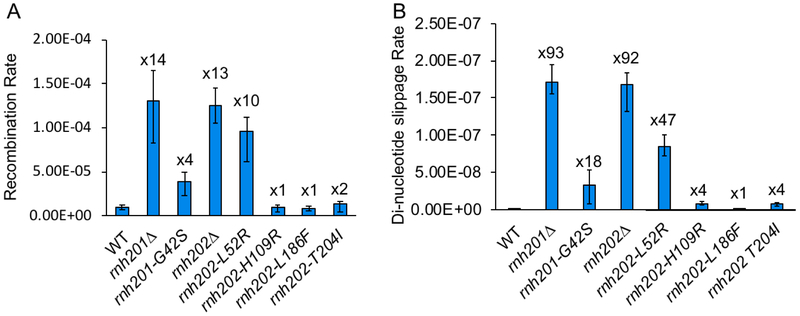
We used intrachromosomal gene conversion and mutation of a slippage repeat reporter to examine the impact of AGS alleles on genome stability. In accordance with the synthetic sickness data, rnh201-G42S and the rnh202-L52R mutants showed an increase in recombination rate, although the level was not as high as the null mutant (Figure 3A). Nonetheless, the increase in recombination in the rnh202-L52R mutant approached that of the null allele and was significantly higher than the rnh201-G42S mutant, indicating a strong loss of RNase H2 function. The rnh202-H109R, rnh202-L186F and rnh202-T204I mutants had recombination levels similar to those of wild type cells (Figure 3A).
Figure 3. Genome instability assays.
A. Recombination rates of rnaseh2-AGS mutants. A recombination reporter consisting of direct LEU2 repeats flanking a URA3 marker was used for fluctuation analysis to determine gene conversion rates. Error bars represent the standard deviation from 3 independent experiments, and fold increase from wild type level is reported above the bars. B. Dinucleotide slippage mutation rate of rnaseh2-AGS mutants. Rates of dinucleotide slippage were determined using a reporter containing an (AG)4 tandem repeat embedded in the LYS2 gene. Fluctuation analysis was used to determine rates, and error bars represent the standard deviation from 3 independent experiments, and fold increase from wild type level is reported above the bars.
We evaluated the mutation using a reporter system that assays specifically for slippages in an (AG)4 repeat sequence [10, 27] as RNase H2 null alleles have a 100X increase in mutation rate, giving a wide range for assessment of hypomorphic mutations. The rnh201-G42S mutant had a significant increase in the dinucleotide slippage rate that was approximately the same fractional increase compared to the null allele rate as that seen for the recombination rate (Figure 3B). Similarly, the mutation slippage rate for the rnh202-L52R mutant was greatly increased, to close to the same fractional increase for the recombination rate (Figure 3B). The greater impact of the rnh202-L52R mutation on genome stability is also in accord with the synthetic lethal growth phenotype with rnh1Δ top1Δ. As expected, the rnh202-H109R, rnh202-L186F and rnh202-T204I mutant strains had dinucleotide slippage rates similar to those of wildtype (Figure 3B).
3.4. Yeast AGS alleles and ribonucleotides in DNA
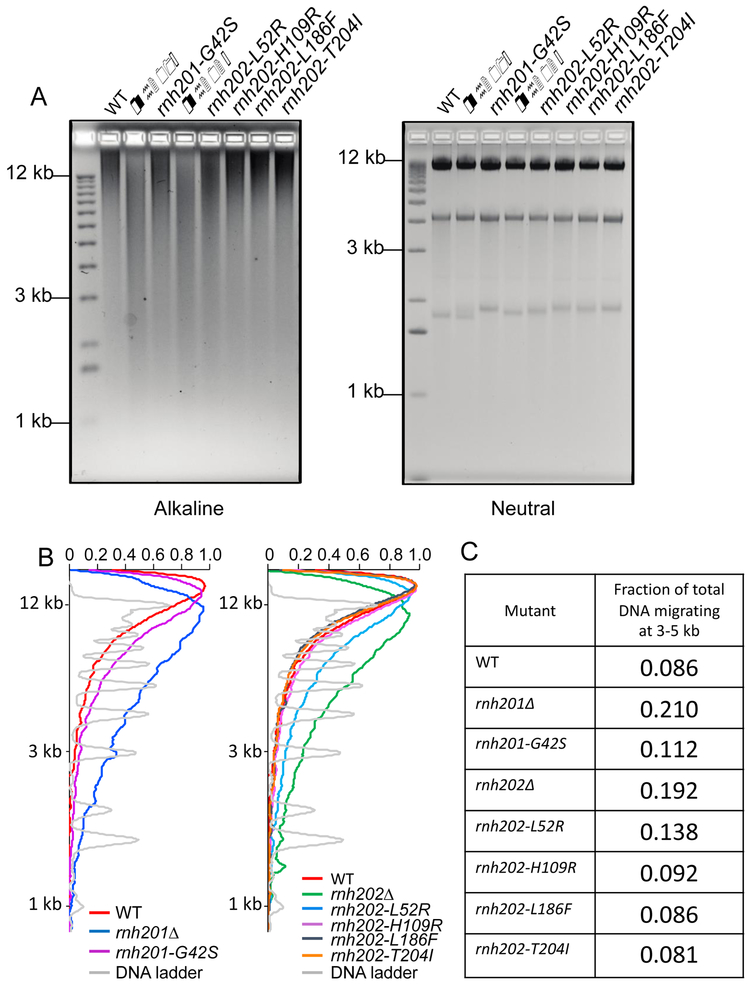
Genomic ribonucleotides can be detected by fragmentation on alkaline gels after alkaline treatment. Deletion of RNH201 or RNH202 results in a marked increase in smearing of DNA on alkaline gels, indicating more ribonucleotides embedded in these genomes (Figure 4). There are no detectable differences on neutral gels. Densitometric tracings of the alkaline gels shows increased fragmentation in the genomic DNA of the rnh201-G42S and rnh202-L52R strains and this is confirmed by determining the fraction of total DNA fragments migrating between 3 kb and 5 kb. Both null strains rnh201Δ and rnh202Δ show an increase in this fraction compared to wildtype following alkaline treatment. In contrast, genomic DNAs from the rnh202-H109R, rnh202-L186F, and rnh202-T204l strains showed no discernable changes in migration compared to RNH202 following alkaline treatment. These results suggest reduced RNase H2 activity in these mutant strains, which most likely accounts for the increased mutation and recombination rates and synthetic phenotypes with rad50Δ and rnh1Δ top1Δ mutations.
Figure 4. Assessment of ribonucleotides in genomic DNA.
A. Alkaline gel analysis of genomic DNA. Genomic DNA was isolated from the indicated strains, treated with 0.3 N KOH for 2 hours at 55°C and run on 1% agarose gel in alkaline buffer. Gels were neutralized and stained with ethidium bromide. For the neutral gel, untreated DNA was run on 1% agarose gel in 1XTBE and stained with ethidium bromide. B. Scans of alkaline gel lanes. Densitometry tracings using MatLab of lanes of the alkaline gel in (A), presenting the RNH201 alleles and the RNH202 alleles separately. C. Fraction of total DNA fragments migrating at 3-5 kb in each lane. The fraction of fragments migrating at 3-5 kb in each sample on the alkaline gel was calculated from the MatLab tracings. Genomic DNA from the rnh201-G42S and rnh202-L52R strains show an increase in this fraction compared to wildtype but in both cases it is less than the respective null strain. The X-axis has arbitrary labels, normalizing the maximum of each peak to 1.
4. Discussion and conclusions
The allele rnh201-G42S in the RNASEH2A/RNH201 gene encodes a variant protein that is significantly reduced in enzymatic activity [16, 18, 19], Our assays for genome instability and DNA migration on alkaline gels show that the rnh201-G42S mutant has some phenotypes indistinguishable from the null allele but others that lie between wildtype and the null allele. Previous studies showed that the rnh201-G42S allele has a mutational profile of the CAN1 gene that is altered from the wildtype allele but is not as strong as the rnh201Δ null allele [13], Alkaline gel analysis suggested a slight increase in fragment mobility of DNA from the rnh201-G42S strain. In contrast, the rnh201-G42S rnh1Δ top1Δ strain gave good growth that was barely distinguishable from wildtype, as initially shown [13], perhaps indicating that the residual activity is sufficient for removal of R-loops. This modest effect in yeast should be considered in contrast to the phenotype seen in mouse where the Rnaseh2A-G37S mutation is a perinatal lethal [28], The RNH202 alleles studied here occur as compound heterozygotes in AGS patients and frequently the other allele is RNASEH2B (c.529G>A, p.Ala177Thr) [2, 16], We observed a range of phenotypes among the rnh202 strains that we generated. The fact that the mutations give rise to AGS when the other allele is RNASEH2B (c.529G>A, p.Ala177Thr) suggests that there is some impairment of the protein function in the compound heterozygous state. As the human genome is approximately 100-fold the size of the S. cerevisiae genome, with approximately 100-fold more rNMP residues incorporated into the genome every replication cycle [6, 9, 29], hypomorphic RNASEH2B mutations may have a phenotypic consequence in humans not observable in S. cerevisiae. Some mutant AGS proteins have normal activity against single rNMP residues in DNA but are defective against R-loops, another substrate for the RNase H2 activity.
In contrast, the mh202-L52R/RNASEH2B (c.179T>G, p.Leu60Arg) allele showed significant defects in all assays, suggesting that rNMP residues are not efficiently removed by the mutant RNase H2 protein and further processing contributes to genome instability and DNA damage. However, the S. cerevisiae RNase H2 protein with the Rnh202L52R subunit has 74% of wildtype activity in vitro [18]. The mutant RNase H2 complex could be more unstable in vivo or in lower amounts, or affected in recruitment to rNMP substrates in vivo. Whether the AGS phenotypes are more severae in patients with this allele and whether this contributes to the disease presentation is not known.
The discordance between the in vitro biochemistry and the in vivo yeast phenotypes and disease presentation in humans remains a challenge to understand the causes of AGS disease. The cGAS/STING pathway is stimulated by DNA damage [30, 31] and RNase H2 AGS mutations can stimulate the cGAS/STING pathway [28, 32]. Still, a direct link between DNA damage, genome instability and AGS presentation is not proven. The DNA damage sensor MRE11, which interacts with RAD50, is conserved between S. cerevisiae and humans and in human cells it is linked to the cGAS/STING pathway through recognition of cytosolic double-stranded DNA [33]. Recent studies on Topoisomerase I acting at rNMPs in DNA have shown that double strand breaks can be formed [34], and these could be the initiating substrate for homologous recombination. As RNase H2 has two activities, one against single rNMP residues in DNA and the second against R-loops [4, 13], it is possible that some AGS phenotypes arise from altered activity against R-loops. Indeed, cells with AGS alleles have increased R-loop accumulation [7].
Supplementary Material
Highlights.
AGS mutations in RNase H2 subunits can be modeled in yeast
Some AGS alleles have no observable phenotype in yeast
Other AGS alleles have strong negative impact on genome stability
in vivo yeast phenotypes correlate with increased retention of rNMPs in DNA
Acknowledgments
We thank Duncan Smith for comments. Eli Rothenberg and Yandong Yin provided invaluable help with scanning and quantification of the alkaline gels. Beatrix Ueberheide provided assistance with protein alignments. Support from the National Institutes of Health R01CA146940 is acknowledged.
Abbreviations:
- AGS
Aicardi-Goutières Syndrome
- DSB
double-strand break
- HR
homologous recombination
- Top1
topoisomerase 1
- LOH
loss of heterozygosity
- RER
ribonucleotide excision repair
- dNTP
deoxynucleotide triphosphate
- rNTP
ribonucleotide triphosphate
- rNMP
ribonucleotide monophosphate
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Aicardi J, Goutieres F, A progressive familial encephalopathy in infancy with calcifications of the basal ganglia and chronic cerebrospinal fluid lymphocytosis, Ann Neurol, 15 (1984) 49–54. [DOI] [PubMed] [Google Scholar]
- [2].Rice G, Patrick T, Parmar R, Taylor CF, Aeby A, Aicardi J, Artuch R, Montalto SA, Bacino CA, Barroso B, Baxter P, Benko WS, Bergmann C, Bertini E, Biancheri R, Blair EM, Blau N, Bonthron DT, Briggs T, Brueton LA, Brunner HG, Burke CJ, Carr IM, Carvalho DR, Chandler KE, Christen HJ, Corry PC, Cowan FM, Cox H, D'Arrigo S, Dean J, De Laet C, De Praeter C, Dery C, Ferrie CD, Flintoff K, Frints SG, Garcia-Cazorla A, Gener B, Goizet C, Goutieres F, Green AJ, Guet A, Hamel BC, Hayward BE, Heiberg A, Hennekam RC, Husson M, Jackson AP, Jayatunga R, Jiang YH, Kant SG, Kao A, King MD, Kingston HM, Klepper J, van der Knaap MS, Kornberg AJ, Kotzot D, Kratzer W, Lacombe D, Lagae L, Landrieu PG, Lanzi G, Leitch A, Lim MJ, Livingston JH, Lourenco CM, Lyall EG, Lynch SA, Lyons MJ, Marom D, McClure JP, McWilliam R, Melancon SB, Mewasingh LD, Moutard ML, Nischal KK, Ostergaard JR, Prendiville J, Rasmussen M, Rogers RC, Roland D, Rosser EM, Rostasy K, Roubertie A, Sanchis A, Schiffmann R, Scholl-Burgi S, Seal S, Shalev SA, Corcoles CS, Sinha GP, Soler D, Spiegel R, Stephenson JB, Tacke U, Tan TY, Till M, Tolmie JL, Tomlin P, Vagnarelli F, Valente EM, Van Coster RN, Van der Aa N, Vanderver A, Vles JS, Voit T, Wassmer E, Weschke B, Whiteford ML, Willemsen MA, Zankl A, Zuberi SM, Orcesi S, Fazzi E, Lebon P, Crow YJ, Clinical and molecular phenotype of Aicardi-Goutieres syndrome, Am J Hum Genet, 81 (2007) 713–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Crow YJ, Livingston JH, Aicardi-Goutieres syndrome: an important Mendelian mimic of congenital infection, Dev Med Child Neurol, 50 (2008) 410–416. [DOI] [PubMed] [Google Scholar]
- [4].Cerritelli SM, Crouch RJ, Ribonuclease H: the enzymes in eukaryotes, FEBS J, 276 (2009) 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hiller B, Achleitner M, Glage S, Naumann R, Behrendt R, Roers A, Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity, J Exp Med, 209 (2012) 1419–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA, Genome instability due to ribonucleotide incorporation into DNA, Nat Chem Biol, 6 (2010) 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lim YW, Sanz LA, Xu X, Hartono SR, Chedin F, Genome-wide DNA hypomethylation and RNA:DNA hybrid accumulation in Aicardi-Goutieres syndrome, Elife, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte GM, Gornall HL, Oojageer A, Anderson B, Pizzino A, Helman G, Abdel-Hamid MS, Abdel-Salam GM, Ackroyd S, Aeby A, Agosta G, Albin C, Allon-Shalev S, Arellano M, Ariaudo G, Aswani V, Babul-Hirji R, Baildam EM, Bahi-Buisson N, Bailey KM, Barnerias C, Barth M, Battini R, Beresford MW, Bernard G, Bianchi M, Billette de Villemeur T, Blair EM, Bloom M, Burlina AB, Carpanelli ML, Carvalho DR, Castro-Gago M, Cavallini A, Cereda C, Chandler KE, Chitayat DA, Collins AE, Sierra Corcoles C, Cordeiro NJ, Crichiutti G, Dabydeen L, Dale RC, D'Arrigo S, De Goede CG, De Laet C, De Waele LM, Denzler I, Desguerre I, Devriendt K, Di Rocco M, Fahey MC, Fazzi E, Ferrie CD, Figueiredo A, Gener B, Goizet C, Gowrinathan NR, Gowrishankar K, Hanrahan D, Isidor B, Kara B, Khan N, King MD, Kirk EP, Kumar R, Lagae L, Landrieu P, Lauffer H, Laugel V, La Piana R, Lim MJ, Lin JP, Linnankivi T, Mackay MT, Marom DR, Marques Lourenco C, McKee SA, Moroni I, Morton JE, Moutard ML, Murray K, Nabbout R, Nampoothiri S, Nunez-Enamorado N, Oades PJ, Olivieri I, Ostergaard JR, Perez-Duenas B, Prendiville JS, Ramesh V, Rasmussen M, Regal L, Ricci F, Rio M, Rodriguez D, Roubertie A, Salvatici E, Segers KA, Sinha GP, Soler D, Spiegel R, Stodberg TI, Straussberg R, Swoboda KJ, Suri M, Tacke U, Tan TY, te Water Naude J, Wee Teik K, Thomas MM, Till M, Tonduti D, Valente EM, Van Coster RN, van der Knaap MS, Vassallo G, Vijzelaar R, Vogt J, Wallace GB, Wassmer E, Webb HJ, Whitehouse WP, Whitney RN, Zaki MS, Zuberi SM, Livingston JH, Rozenberg F, Lebon P, Vanderver A, Orcesi S, Rice GI, Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1, Am J Med Genet A, 167A (2015) 296–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, Devenney PS, Sexton D, Grimes G, Holt IJ, Hill RE, Taylor MS, Lawson KA, Dorin JR, Jackson AP, Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development, Cell, 149 (2012) 1008–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S, Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I, Science, 332 (2011) 1561–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Potenski CJ, Niu H, Sung P, Klein HL, Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms, Nature, 511 (2014) 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Aguilera A, Klein HL, Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations, Genetics, 119 (1988) 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chon H, Sparks JL, Rychlik M, Nowotny M, Burgers PM, Crouch RJ, Cerritelli SM, RNase H2 roles in genome integrity revealed by unlinking its activities, Nucleic Acids Res, 41 (2013) 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Epshtein A, Potenski CJ, Klein HL, Increased Spontaneous Recombination in RNase H2- Deficient Cells Arises From Multiple Contiguous rNMPs and Not From Single rNMP Residues Incorporated by DNA Polymerase Epsilon, Microb Cell, 3 (2016) 248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ii M, Ii T, Mironova LI, Brill SJ, Epistasis analysis between homologous recombination genes in Saccharomyces cerevisiae identifies multiple repair pathways for Sgs1, Mus81-Mms4 and RNase H2, Mutat Res, 714 (2011) 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, Bertini E, Chandler KE, Chitayat D, Cau D, Dery C, Fazzi E, Goizet C, King MD, Klepper J, Lacombe D, Lanzi G, Lyall H, Martinez-Frias ML, Mathieu M, McKeown C, Monier A, Oade Y, Quarrell OW, Rittey CD, Rogers RC, Sanchis A, Stephenson JB, Tacke U, Till M, Tolmie JL, Tomlin P, Voit T, Weschke B, Woods CG, Lebon P, Bonthron DT, Ponting CP, Jackson AP, Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection, Nat Genet, 38 (2006) 910–916. [DOI] [PubMed] [Google Scholar]
- [17].den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, Roux AF, Smith T, Antonarakis SE, Taschner PE, HGVS Recommendations for the Description of Sequence Variants: 2016 Update, Hum Mutat, 37 (2016) 564–569. [DOI] [PubMed] [Google Scholar]
- [18].Rohman MS, Koga Y, Takano K, Chon H, Crouch RJ, Kanaya S, Effect of the disease-causing mutations identified in human ribonuclease (RNase) H2 on the activities and stabilities of yeast RNase H2 and archaeal RNase HII, FEBS J, 275 (2008) 4836–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chon H, Vassilev A, DePamphilis ML, Zhao Y, Zhang J, Burgers PM, Crouch RJ, Cerritelli SM, Contributions of the two accessory subunits, RNaSeH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex, Nucleic Acids Res, 37 (2009) 96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Figiel M, Chon H, Cerritelli SM, Cybulska M, Crouch RJ, Nowotny M, The structural and biochemical characterization of human RNase H2 complex reveals the molecular basis for substrate recognition and Aicardi-Goutieres syndrome defects, J Biol Chem, 286 (2011) 10540–10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Reijns MA, Bubeck D, Gibson LC, Graham SC, Baillie GS, Jones EY, Jackson AP, The structure of the human RNase H2 complex defines key interaction interfaces relevant to enzyme function and human disease, J Biol Chem, 286 (2011) 10530–10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Spell RM, Jinks-Robertson S, Determination of mitotic recombination rates by fluctuation analysis in Saccharomyces cerevisiae, Methods Mol Biol, 262 (2004) 3–12. [DOI] [PubMed] [Google Scholar]
- [23].Lea DE, Coulson CA, The distribution of the numbers of mutants in bacterial populations, J Genet, 49 (1949) 264–285. [DOI] [PubMed] [Google Scholar]
- [24].Roy A, Kucukural A, Zhang Y, I-TASSER: a unified platform for automated protein structure and function prediction, Nat Protoc, 5 (2010) 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Roy A, Yang J, Zhang Y, COFACTOR: an accurate comparative algorithm for structure-based protein function annotation, Nucleic Acids Res, 40 (2012) W471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].El Hage A, French SL, Beyer AL, Tollervey D, Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis, Genes & development, 24 (2010) 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lippert MJ, Kim N, Cho JE, Larson RP, Schoenly NE, O'Shea SH, Jinks-Robertson S, Role for topoisomerase 1 in transcription-associated mutagenesis in yeast, Proc Natl Acad Sci U S A, 108 (2011) 698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pokatayev V, Hasin N, Chon H, Cerritelli SM, Sakhuja K, Ward JM, Morris HD, Yan N, Crouch RJ, RNase H2 catalytic core Aicardi-Goutieres syndrome-related mutant invokes cGASSTING innate immune-sensing pathway in mice, J Exp Med, 213 (2016) 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA, Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases, Proc Natl Acad Sci U S A, 107 (2010) 4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA, Mitotic progression following DNA damage enables pattern recognition within micronuclei, Nature, 548 (2017) 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, Olova N, Sutcliffe H, Rainger JK, Leitch A, Osborn RT, Wheeler AP, Nowotny M, Gilbert N, Chandra T, Reijns MAM, Jackson AP, cGAS surveillance of micronuclei links genome instability to innate immunity, Nature, 548 (2017) 461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mackenzie KJ, Carroll P, Lettice L, Tarnauskaite Z, Reddy K, Dix F, Revuelta A, Abbondati E, Rigby RE, Rabe B, Kilanowski F, Grimes G, Fluteau A, Devenney PS, Hill RE, Reijns MA, Jackson AP, Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response, EMBO J, 35 (2016) 831–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, Komatsu K, Akira S, Kawai T, DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking, Proc Natl Acad Sci U S A, 110 (2013) 2969–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Huang SN, Williams JS, Arana ME, Kunkel TA, Pommier Y, Topoisomerase I-mediated cleavage at unrepaired ribonucleotides generates DNA double-strand breaks, EMBO J, 36 (2017) 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.