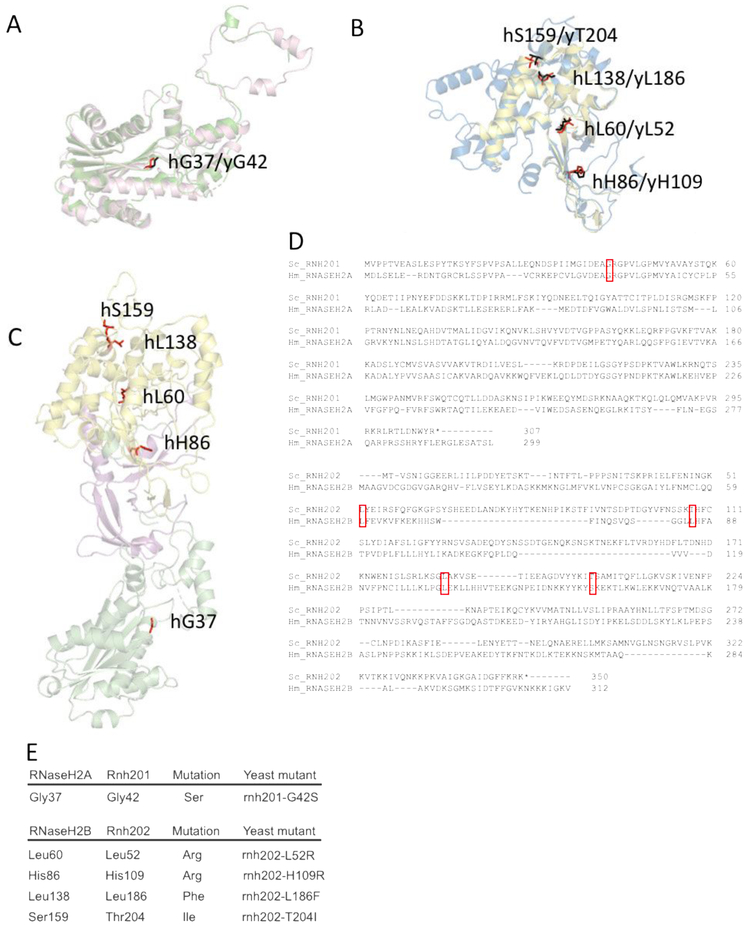
Figure 1. Conserved AGS-associated residues from yeast to humans.
Homology models of yeast Rnh201 and Rnh202 were generated and aligned to structural models of human RNASEH2A and RNASEH2B, respectively. Conserved AGS-associated residues were identified on the aligned yeast homology structures. A. We chose one conserved residue in yRnh201 (magenta) / hRNASEH2A (green): yGly42(red) /hGly37(black). B. We chose four conserved residues in yRnh202(blue) / hRNASEH2B(yellow): 1.) yHis109/hHis86; 2.) yLeu52/hLeu60; 3.) yLeu186/hLeu138; 4.) yThr204/hSer159 (yeast residues in red, human residues in black). C. Conserved residues identified are located in different parts of the enzyme complex. The structure (PDB ID = 3PUFR) of the RNase H2 heterotrimeric complex consisting of RNASEH2A (green), RNASEH2B (yellow) and RNASEH2C (magenta) is shown with AGS-associated residues (red) that are conserved in yeast highlighted. D. Alignment of yeast Rnh201 with human RNASEH2A and yeast Rnh202 with human RNASEH2B. The top panel shows the alignment of Rnh201 with RNASEH2A using Clustal Omega. The lower panel shows the alignment of Rnh202 with RNASEH2B using Clustal Omega. The red boxes indicate the position of the human AGS mutations shown in panels A, B and E and their alignment with the cognate yeast residue. E. Mutations of the RNase H2 complex found in AGS patients are reconstructed in yeast strains. Yeast cells with the appropriate amino acid change that reflects AGS mutations were constructed.