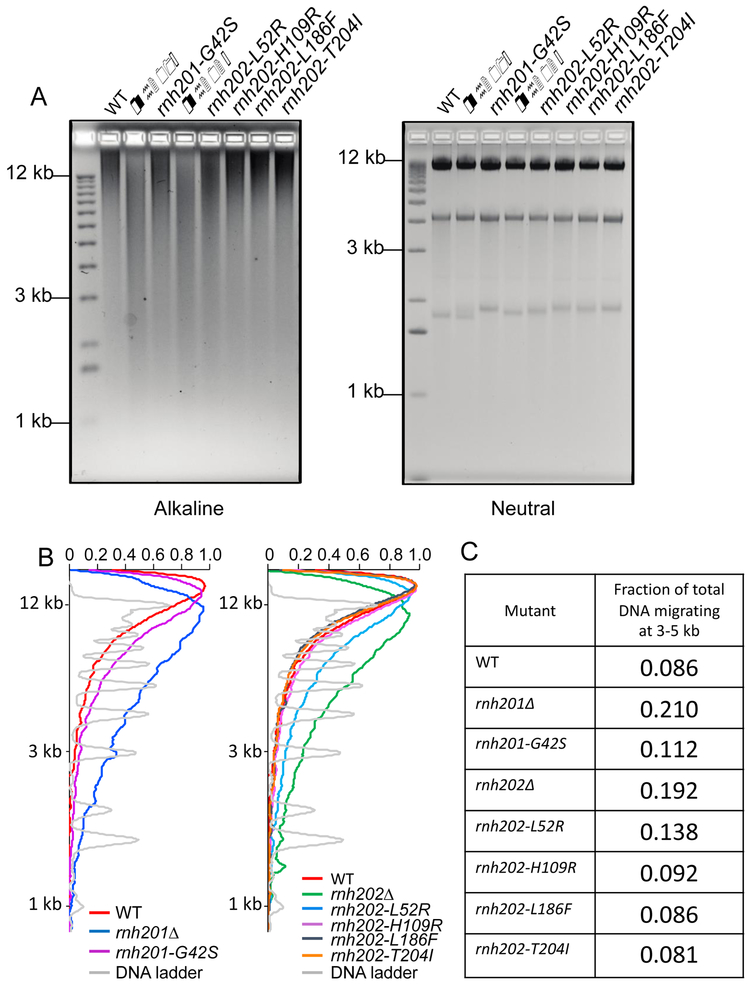
Figure 4. Assessment of ribonucleotides in genomic DNA.
A. Alkaline gel analysis of genomic DNA. Genomic DNA was isolated from the indicated strains, treated with 0.3 N KOH for 2 hours at 55°C and run on 1% agarose gel in alkaline buffer. Gels were neutralized and stained with ethidium bromide. For the neutral gel, untreated DNA was run on 1% agarose gel in 1XTBE and stained with ethidium bromide. B. Scans of alkaline gel lanes. Densitometry tracings using MatLab of lanes of the alkaline gel in (A), presenting the RNH201 alleles and the RNH202 alleles separately. C. Fraction of total DNA fragments migrating at 3-5 kb in each lane. The fraction of fragments migrating at 3-5 kb in each sample on the alkaline gel was calculated from the MatLab tracings. Genomic DNA from the rnh201-G42S and rnh202-L52R strains show an increase in this fraction compared to wildtype but in both cases it is less than the respective null strain. The X-axis has arbitrary labels, normalizing the maximum of each peak to 1.