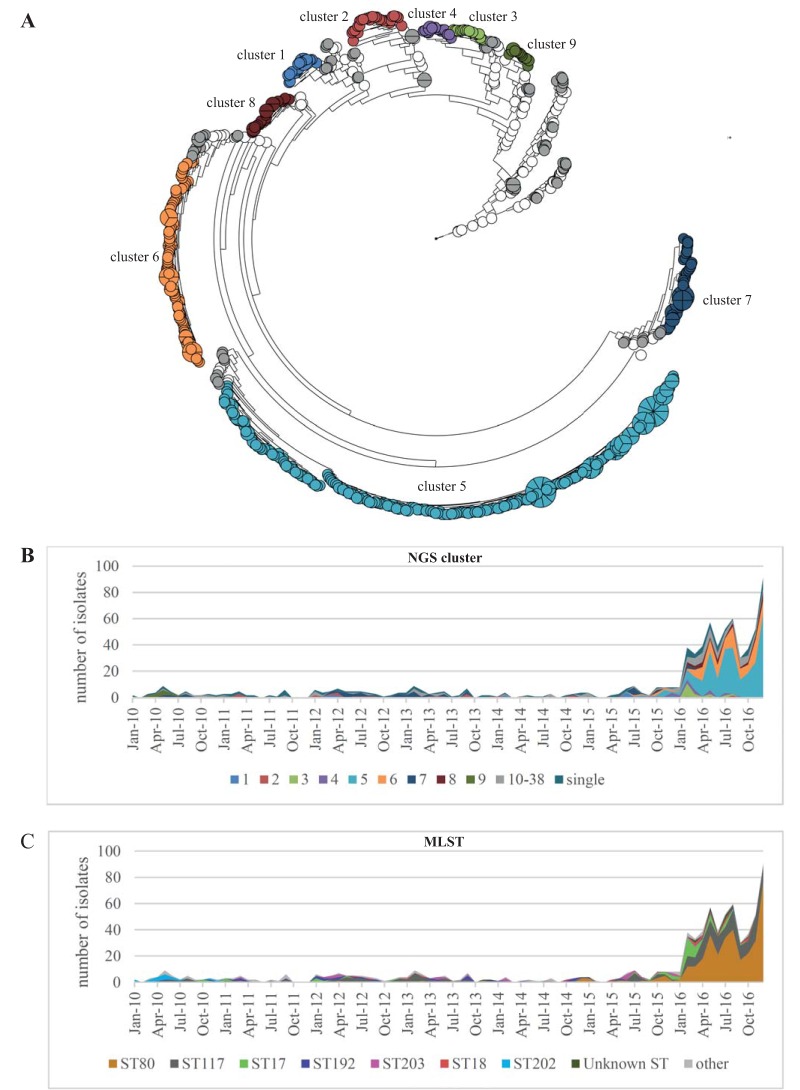
FIG 1.
(A) Phylogeny of 773 VREfm study isolates based on pairwise comparison of core genome SNP similarities and clustering using unweighted pair group method using average linkages (UPGMA). The branch length is shown on a logarithmic scale. In total, 38 NGS clusters were defined based on SNP differences of the isolates. NGS clusters 1 to 9 comprised more than 10 isolates each and are displayed in different colors, whereas the NGS clusters 10 to 38 with fewer than 10 isolates are displayed in gray. White circles represent singletons. (B and C) Temporal distribution of NGS clusters and MLST of the isolates over time. The increase of VREfm cases during the outbreak was mainly due to the expansion of isolates of NGS clusters 5 and 6. Interestingly, NGS cluster 5 and cluster 6 were not present in our hospital until June and July 2016, respectively, shortly before the outbreak started.