Figure 1.
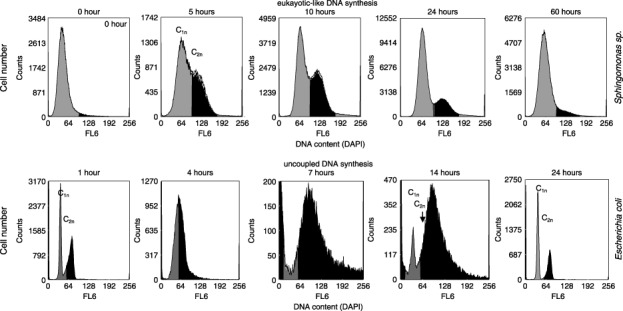
Eukaryotic‐like DNA synthesis: flow cytometric analysis of the DNA content of Gram‐negative Sphingomonas sp. LB126 cultivated for 60 h. The species was grown aerobically at 20 °C and pH 7.5 in 500 mL shake flasks with 300 mL minimal medium at 150 r.p.m. The carbon and energy source was glucose (1 g/L). The harvested cells were centrifuged at 3200 g for 5 min, fixed with 10% NaN3, and stored at 4 °C. Bacterial DNA was stained by treatment of 2 mL of diluted cell suspension (3 × 108 cells per millilitre) with 1 mL solution A (2.1 g citric acid/0.5 g Tween 20 in 100 mL bidistilled water) for 10 min. Afterwards the cells were washed and re‐suspended in 2 mL solution B (0.24 µm 4′,6‐diamidino‐2‐phenylindole [DAPI, SIGMA], 400 mm Na2HPO4, pH 7.0) for at least 20 min (according to Shi et al. 2007). Flow cytometric measurements were carried out using a MoFlo cell sorter (DakoCytomation, Fort Collins, CO, USA) equipped with two water‐cooled argon‐ion lasers (Innova 90C and Innova 70C from Coherent, Santa Clara, CA, USA). Chromosome equivalents are presented as C1n and C2n. Uncoupled DNA synthesis: The Gram‐negative Escherichia coli K12 was grown aerobically at 30 °C and pH 7.5 for 24 h in 500 mL shake flasks at 150 r.p.m. on 300 mL peptone media (L): 5 g peptone from meat (pancreatic), 3 g NaCl, 2 g K2HPO4, 10 g meat extract, 10 g yeast extract, 5 g glucose. The staining procedure and device equipment was the same as above though the DAPI concentration was enhanced to 0.5 µm DAPI within solution B. Chromosome equivalents are presented as C1n and C2n. Uncoupled DNA synthesis is shown to be performed between the 7th and 14th hours.
