Figure 2.
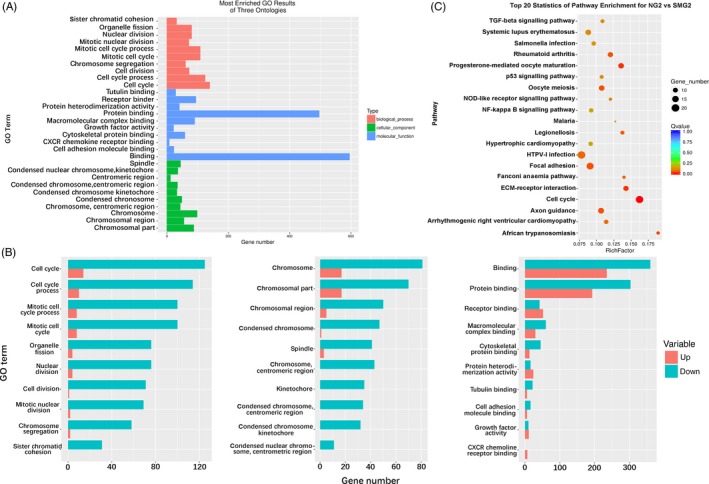
GO analysis of DEGs in three ontologies and Pathway analysis of DEGs in NG2 vs SMG2 samples. (A) Red, blue and green represent biological process, cellular component and molecular function, respectively. (B) Up‐ and down‐regulated genes enriched for three ontologies, biological process, molecular function and cellular component from left to right, respectively. Red represents up‐regulated genes, and green represents down‐regulated genes. (C) The size of dot represents the number of DEGs. Rich Factor refers to the ratio between DEGs enriched in this pathway and all the annotated genes in this pathway. A large enrichment factor denotes a high degree of enrichment. The lower the q‐value, the more significant the enrichment of DEGs. GO, gene ontology; DEGs, differentially expressed genes; NG2, cells induced for 2 d under normal ground condition; SMG2, cells induced for 2 d under simulated microgravity
