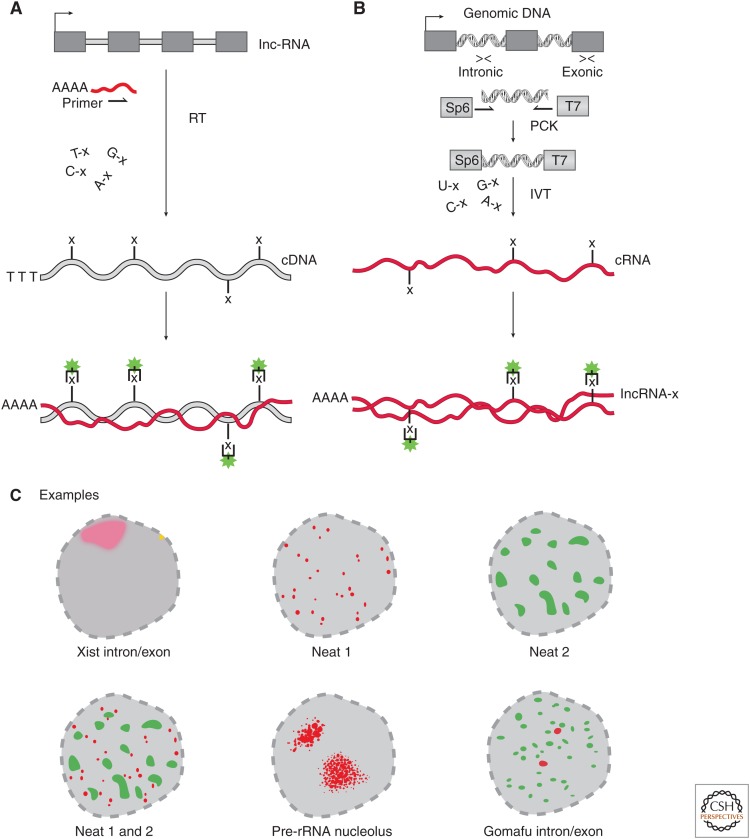
Figure 1.
Classic RNA fluorescence in situ hybridization (RNA-FISH) probe design using complementary DNA (cDNA) and complementary RNA (cRNA) probes. (A) To generate cDNA probes for RNA-FISH, one performs reverse transcription (RT) with gene-specific primers to generate a cDNA to the targeted long noncoding RNA (lncRNA). During the RT reaction, nucleotides are randomly incorporated that have modifications (e.g., “x” = biotin or digoxigenin [DIG]). The labeled cDNA is hybridized to the endogenous lncRNA (red line), which can be imaged via addition of fluorescent secondary conjugates targeting the probe label. (B) Generation of cRNA RNA-FISH probes requires targeted polymerase chain reaction (PCR) of genomic DNA, targeting either the intronic or exonic regions of the lncRNA locus. PCR primers have 5′/3′ sequences appended encoding in vitro transcription (IVT) promoters that are incorporated into the final double-stranded DNA (dsDNA) product. The resulting dsDNA product is subjected to IVT either antisense (complementary) or antisense (noncomplementary) in the presence of modified nucleotides (same as in A) to produce a labeled cRNA probe (red line with “x”). The cRNA probe is then hybridized to the endogenous lncRNA target, which can be imaged via addition of fluorescent secondary conjugates targeting the probe label. (C) lncRNA-FISH examples of classic studies revealing new subnuclear structures (all images showing nucleus). (Top left) Xist showing intron and exon labeling of the Xist lncRNA showing a punctate site of transcription (yellow) and mature transcript as a “cloud” in red on the same chromosome as the site of transcription. (Top middle and right, bottom left) Cartoon rendition of the RNA-FISH nuclear localization pattern for Neat1, Neat2, and Neat1/2, respectively, with nonoverlapping “speckled” localization patterns. (Bottom middle) rRNA-FISH demarcates the nucleolus, the site of rRNA transcription. (Bottom right) Gomafu RNA-FISH with intronic (red) and exonic (green) RNA-FISH probes shows that the mature transcript localizes in large domains away from the site of transcription (intronic signal).