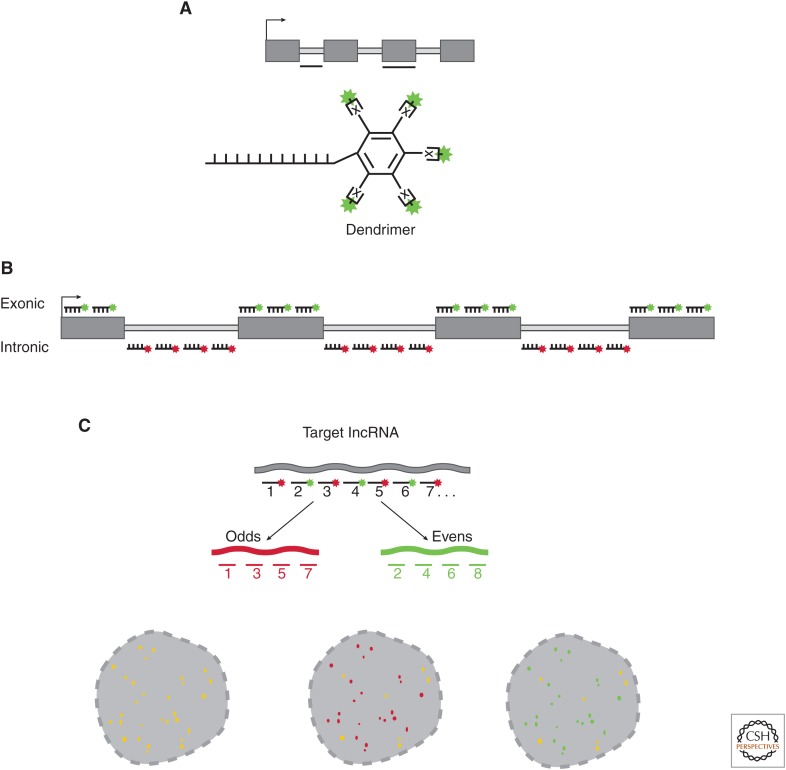
Figure 2.
Probe design for single-molecule RNA fluorescence in situ hybridization (smRNA-FISH). (A) The dendrimer or branched oligo approach. Oligo(s) are designed to specify an intronic or exonic region of the target long noncoding RNA (lncRNA). The dendrimer approach appends either multiple conjugation sites (x) to the 5′ or 3′ end of the oligo. Alternatively, “branched-oligos” are similar to dendrimers using multiple “branched” sequences that form a tree-like structure of conjugatable epitopes. (B) smRNA-FISH probe design using “oligo pools.” Each oligo ranges in length between 18 and 22 nucleotides (nt), and between 20 and 48 oligos are mixed into an oligo pool. Each oligo has modifications on the 5′ or 3′ end to allow conjugation of a fluorophore of choice. smRNA-FISH oligo pools are generated targeting either the intronic regions (site of lncRNA transcription) or exonic regions of the mature transcript. (C) smRNA-FISH probe validation. As a first test for oligo specificity, the pool can be split evenly and each half labeled with distinct fluorophores. If all oligos from the odd (red) and even (green) probe sets are specific to the targeted long noncoding RNA (lncRNA), the patterns will overlap producing yellow signal (left). If one or more of the odd (red) oligos hybridize to an off-target RNA, but most of the probes are specific, then yellow and red signal would be observed (middle). Similarly, if one of the even (green) probes had an off-target RNA, but some probes were specific, green and yellow signal would be observed (right).