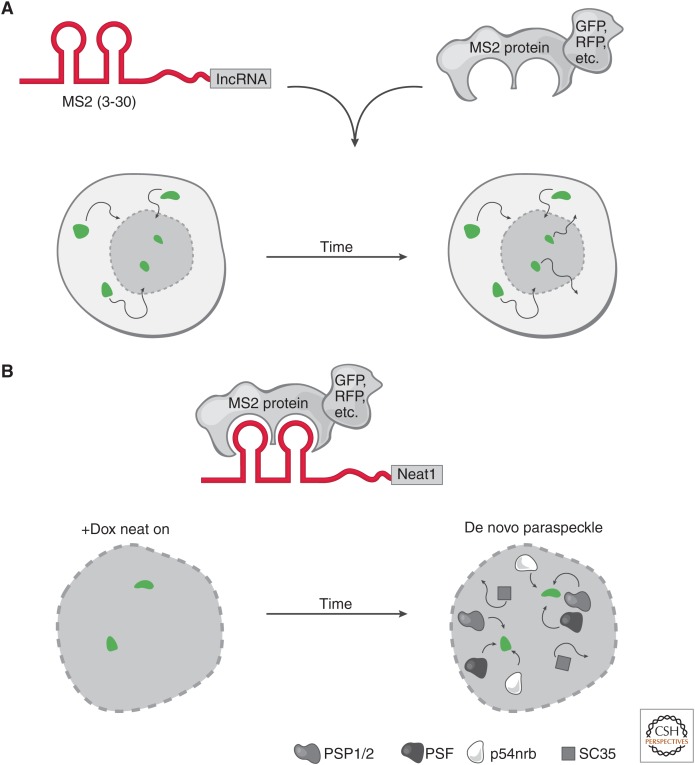
Figure 3.
Live-cell RNA imaging. (A) The targeted long noncoding RNA (lncRNA) has a specific RNA sequence/structure (aptamer) appended (e.g., a series of MS2 hairpins). In parallel, a protein that specifically and tightly binds to the RNA aptamer is fused to a fluorescent marker. These constructs can be integrated at the endogenous locus or ectopically expressed in cells. Conjugation of the RNA aptamer with the cognate fusion protein provides the ability to track the fluorescent signal in live cells through time. This highlights the movements (arrows) of a long noncoding RNA (lncRNA), such as its rate of nuclear and cytoplasmic transport (bottom nuclei). (B) Neat1 live-cell RNA imaging. Live-cell RNA imaging of Neat1 revealed key insights into the role of its transcription in the establishment and maintenance of nuclear paraspeckles. The Neat1 RNA was tracked by appending 24× MS2 hairpins and cognate MS2 protein fused to a fluorescent protein. Neat1 expression was induced (green foci) by doxycycline (+Dox) and over time, paraspeckle proteins (PSP1/2, PSF, p54nrb) were recruited to the de novo expression sites of Neat1 RNA. In contrast, nuclear speckle protein (SC35) was not recruited to the de novo paraspeckle. When Neat1 expression was turned off in the absence of Dox, the paraspeckle disassembles. Together, these observations show that expression of Neat1 precedes paraspeckle formation and is required to maintain paraspeckles in the nucleus.