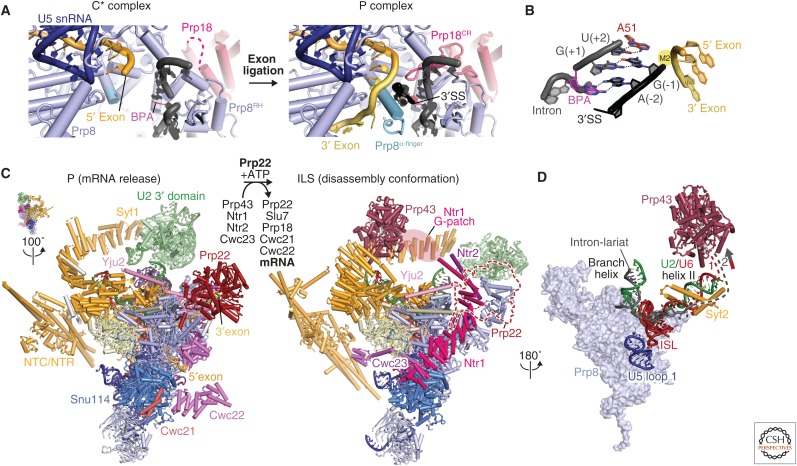
Figure 7.
Exon ligation and spliceosome disassembly. (A) Comparison of the C* and P complex active sites. The extended Prp8 α-finger (light blue) and the Prp18 conserved region (CR) (pink) stabilize 3′SS docking in the active site. This positions the 3′-OH group of the 5′ exon to attack at the 3′SS for exon ligation (PDB 6EXN). (B) Recognition of the conserved YAG sequence at the 3′SS. The last intron nucleotide, G(−1), forms a non-Watson–Crick base pair with the first intron nucleotide G(+1) and the penultimate intron nucleotide A(–2) forms a symmetric Hoogsteen–Hoogsteen base pair with the branch point adenosine (PDB 6EXN). The third last pyrimidine (Y) is recognized by Gln-1594 of Prp8. (C) Transition from the P (PDB 6EXN) to ILS (intron lariat spliceosome) (PDB 5Y88). Prp22-mediated ligated-exon release facilitates the binding of Prp43/Ntr1/Ntr2/Cwc23 and the release of the step 2 factors (Prp18 and Slu7) and Cwc21 and Cwc22, which interact with 5′ exon. (D) Prp43 induces disassembly of ILS by pulling on either the intron or U6 small nuclear RNA (snRNA) (PDB 5Y88).