Figure 2.
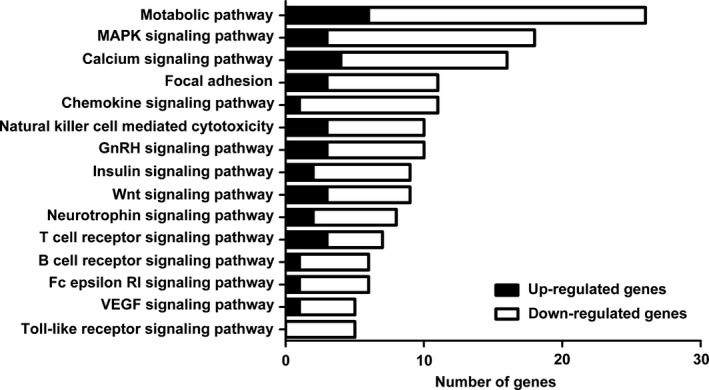
Proportion of differentially expressed genes in significantly affected pathways. KEGG pathway analysis showed that the 15 most related pathways to DEGs included metabolic, mitogen‐activated protein kinase, calcium, focal adhesion, chemokine, natural killer cell‐mediated cytotoxicity, gonadotropin‐releasing hormone, insulin, Wnt, neurotrophin, T‐cell receptor, B cell receptor, Fc epsilon RI, vascular endothelial growth factor and the Toll‐like receptor signalling pathway. Almost all of these pathways were significantly down‐regulated in CD8+ T cells cultured with SKOV3 cells compared with CD8+ T cells cultured alone
