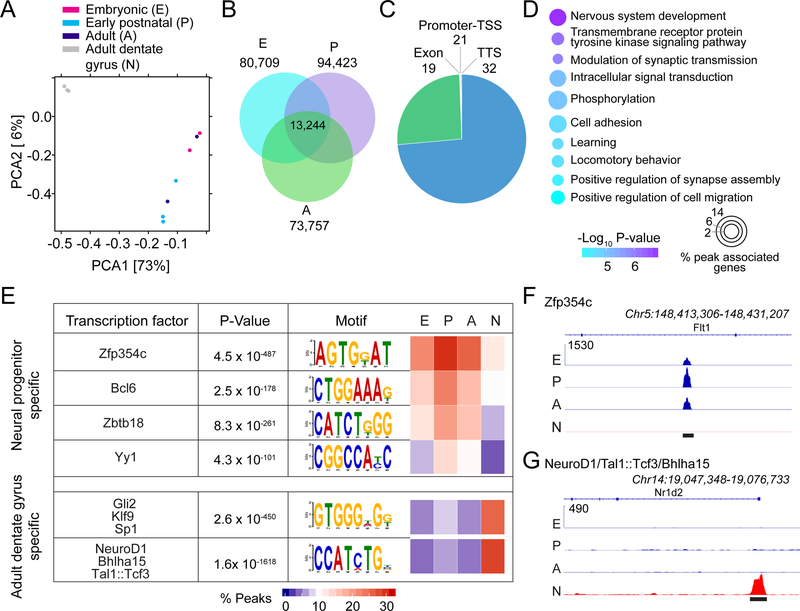
Figure 6. Dentate Hopx+ neural progenitors maintain a stable landscape of chromatin accessibility across development.
(A) PCA plot of ATAC-seq biological replicates of embryonic (E), early postnatal (P), and adult (A) dentate neural progenitors and adult dentate gyrus samples (N). ATAC-seq data for adult dentate gyrus is from (Su et al., 2017).
(B) Venn diagram illustrating the overlap of ATAC-seq peaks in dentate progenitors at different stages.
(C) Genome annotation of the dentate progenitor-enriched peaks, which were determined by comparing common peaks found in neural progenitors (B) to those in the adult dentate gyrus samples. TSS: transcription start site, TTS: transcription termination site.
(D) GO analysis of genes associated with progenitor enriched ATAC-seq peaks in the promoter-TSS, exon, intron, and TTS regions.
(E) Motif discovery analysis of dentate neural progenitor-enriched peaks or adult dentate gyrus-enriched peaks. Motifs shown are known transcription factor binding sites whose transcription factors were expressed in our samples and had an enrichment p-value ≤ 1×10−100.
(F-G) Representative chromatin profiling coverage of dentate neural progenitor-enriched peak with a Zfp354c binding site motif (F), and an adult dentate gyrus-enriched peak with a Neurod1/Tal1:Tcf3/Bhlha15 binding site motif (G). Y-axis indicates normalized reads. Black bars indicate peak locations.