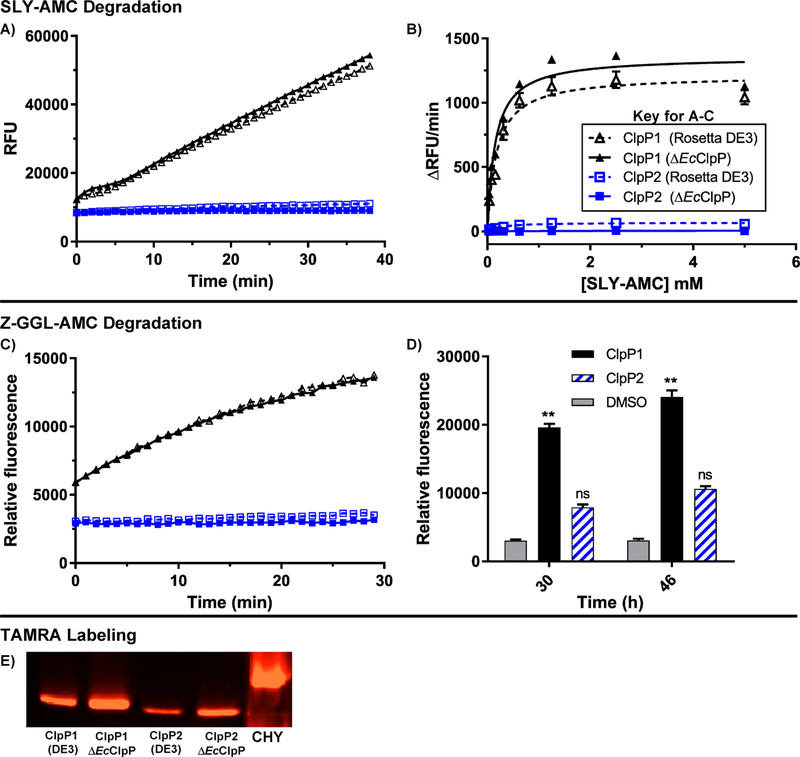
Figure 5.
Peptidolytic activity of ClpP1 and ClpP2. (A) Time-dependent degradation of SLY-AMC by ClpP1 expressed from ΔEcClpP cells (black, solid, solid triangle), ClpP1 from Rosetta DE3 (black, dashed, hollow triangle), ClpP2 from ΔEcClpP (blue, solid, solid square), and ClpP2 from Rosetta DE3 (blue, dashed, hollow square). Experiments conducted with 1 μM ClpP tetradecamer and 0.5 mM SLY-AMC. (B) Michaelis-Menten analysis of ClpP1 and ClpP2 degradation of SLY-AMC. ClpP1 (DE3) Vmax = 1213 ± 38.0; ClpP1 (ΔEcClpP) Vmax = 1357 ± 39.9. ClpP2 Vmax values were not determined due to relatively negligible cleavage of the substrate. (C) Time-dependent (0–30 min) degradation of Z-GGL-AMC by ClpP isoforms. Experiments conducted with 1 μM ClpP tetradecamer and 0.1 mM Z-GGL-AMC. (D) Degradation of Z-GGL-AMC by ClpP isoforms after extended periods (30 and 46 h) of incubation. Means of each data set were compared to the control (Z-GGL-AMC alone) at each time point for statistical significance within a 95% confidence interval by two-way ANOVA analysis with Dunnett’s multiple comparison test. (**, P < 0.05). (E) Serine-protease active site labeling with ActivX TAMRA-FP visualized in an SDS-PAGE gel with chymotrypsin (CHY) as a positive control.