Figure1: Molecular Organization of the Mammalian Circadian Clock.
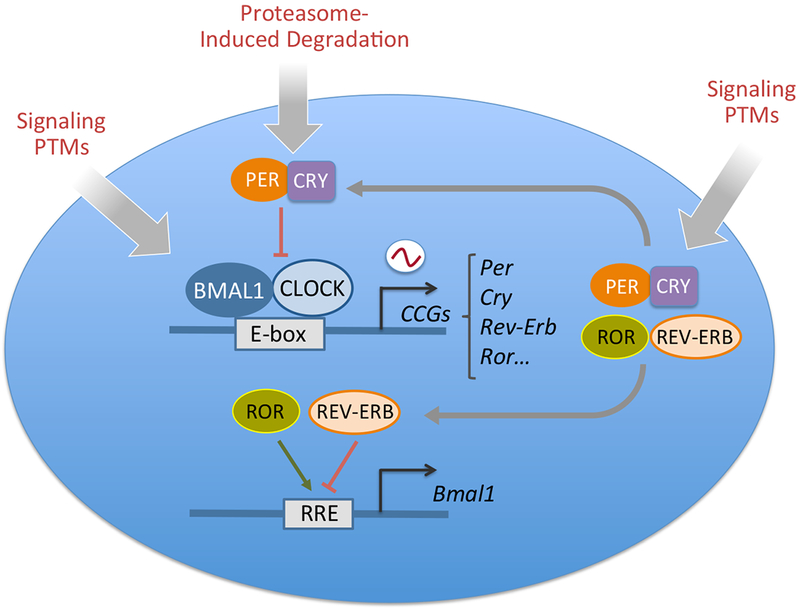
The mammalian molecular clock consists of a positive loop driven by the transcriptional activators CLOCK and BMAL1 and a negative feedback loop driven by the repressors period (PER) and cryptochrome (CRY) proteins. In mammals there are three PER proteins and two CRYs. CLOCK and BMAL1 activate the expression of clock-controlled genes (CCGs) through binding to E-box elements in their promoters. Among the CCGs are Per and Cry genes whose products dimerize and translocate into the nucleus where they inhibit CLOCK:BMAL1 activity. PERs and CRYs undergo a number of post-translational modifications that result in proteasome-induced degradation with a 24 hour rhythmicity, ultimately allowing the start of a new circadian cycle. CLOCK:BMAL1 also induce the activation of Rev-Erb and Ror genes that give rise to a secondary loop by binding to responsive promoter elements (RRE ) and inhibit and activate respectively Bmal1 transcription. Most of the molecular clock components are additionally regulated through various signaling pathways that post-translationally modify the core clock. Post-translational modifications (PTMs) include acetylation, phosphorylation, O-GlcNAcylation and SUMOylation (See Ref 181 for an overview). Together these transcriptional-translational regulatory loops generate the circadian output.
 indicates oscillation.
indicates oscillation.
