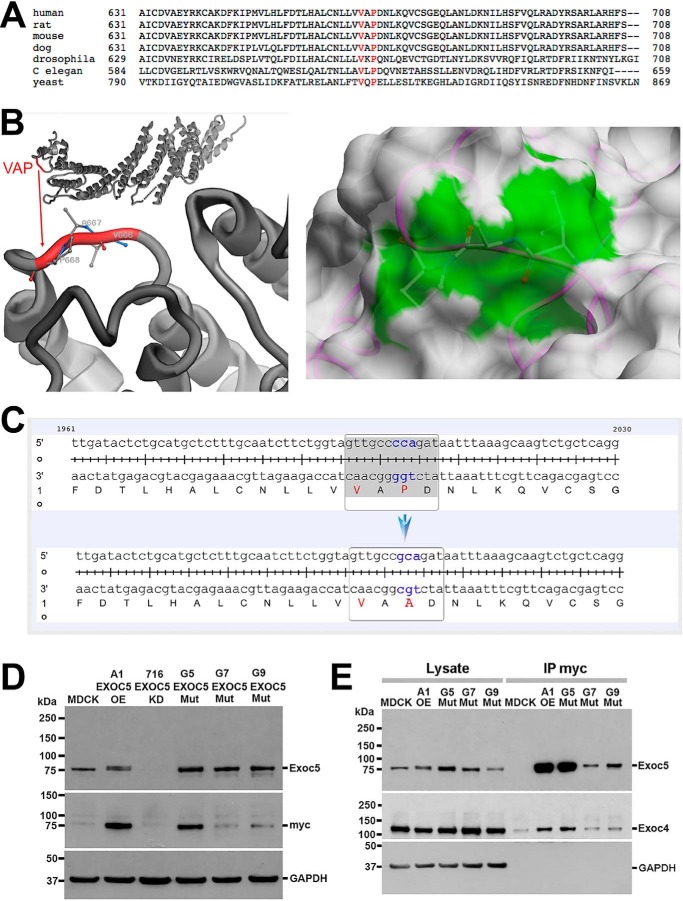
Figure 1.
Site-directed mutagenesis of the ciliary targeting sequence in human EXOC5 cDNA results in a stable protein that can bind other exocyst components. A, the EXOC5 VXPX ciliary targeting sequence is highly conserved from yeast to humans. B, the VXPX ciliary targeting sequence in the EXOC5 3D protein model shows that proline (and to a lesser degree valine) are on the outside of the EXOC5 protein and hence are available for binding. The right panel demonstrates the solvent-accessible surface of EXOC5 in the 5h11 structure. The protein is shown as backbone trace (magenta) and the molecular surface obtained with a spherical water probe with a radius of 1.4 Å (white). Three residues Val666, Ala667, and Pro668 (VAP) are shown by balls and sticks. The contribution of these residues to the molecular surface is marked in green. C, site-directed mutagenesis of cytosine at position 2002 results in a guanine substitution (cca to gca), which leads to alanine being translated instead of a proline. D, EXOC5CTS-m protein is stable as determined by Western blotting. Indeed, lysates from the three stable clonal EXOC5CTS-m cell lines (G5, G7, G9) show more mutated EXOC5 protein than endogenous Exoc5 protein found in control MDCK cells. The amount of mutated EXOC5 protein, especially in clone G5, is similar to the amount of EXOC5 protein that we found in EXOC5 OE cells that we previously generated (clone A1). The mutated and control EXOC5 proteins likely run slower on the gel because of the additional amino acids found in the myc epitope tag. Confirmation of the presence of human EXOC5 protein is demonstrated by staining using 9E10 antibody against the myc epitope tag. E, immunoprecipitation using antibody against the myc epitope tag of the EXOC5CTS-m protein shows that EXOC5CTS-m co-immunoprecipitates endogenous Exoc4. IP, immunoprecipitation; Mut, mutant.