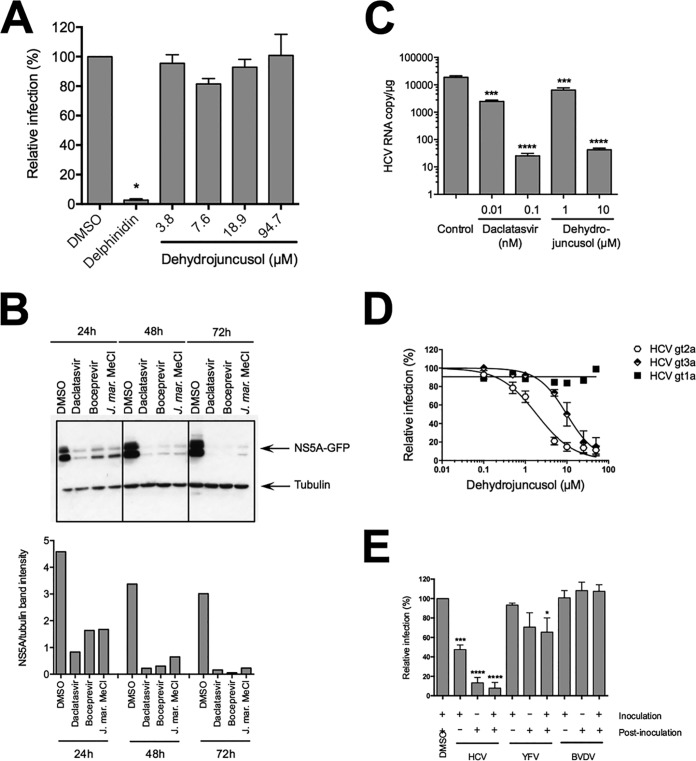
FIG 3.
Dehydrojuncusol inhibits HCV RNA replication. (A) HCVpp were used to inoculate Huh-7 cells in the presence of dehydrojuncusol at indicated concentrations. Delphinidin at 50 µM was used as a control. Cells were lysed 48 h postinoculation and luciferase activity quantified. Data are expressed as a ratio to the DMSO (0.0001%) control (100%). (B) Huh-7 cells stably expressing the SGR-JFH1-NS5AGFP replicon were incubated for the indicated time with either 0.5 nM daclatasvir, 1 µM boceprevir, or 10 µg/ml of the methylene chloride partition of J. maritimus (J. mar. MeCl). DMSO (0.0001%) was used as a control. Cells were lysed and lysates subjected to SDS-PAGE followed by a Western blot revelation of GFP and tubulin. The quantification of band intensity is shown in the graph. (C) HCVcc-infected Huh-7 cells were maintained in the presence of dehydrojuncusol or daclatasvir at the indicated concentrations for 48 h. Cells were lysed, RNA extracted, and HCV genomic RNA quantified by qRT-PCR. Data are representative of three experiments. Error bars represent standard deviation (SD). (D) Huh-7 cells were inoculated with HCVcc of genotype 2a (strain JFH1), genotype 1a (strain TN), and genotype 3a (DBN3a) for 2 h. Inoculum was removed and replaced with a medium containing an increasing concentration of dehydrojuncusol. Cells were fixed 30 h postinfection and subjected to immunofluorescence staining with monoclonal anti-NS5A antibody (9E10). Data are means of values obtained in 3 independent experiments performed in triplicate. Error bars represent SEM. (E) Huh-7 cells were inoculated with HCVcc or YFV and MDBK cells with BVDV in the presence (+) or absence (−) of 10 µg/ml of the methylene chloride partition of J. maritimus. Inoculum was removed and replaced with medium with (+) or without (−) 10 µg/ml of the methylene chloride partition of J. maritimus. Cells were fixed and infection detected by immunofluorescence labeling of viral proteins. Data are means of values obtained in 3 independent experiments performed in triplicate. Error bars represent SEM. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.