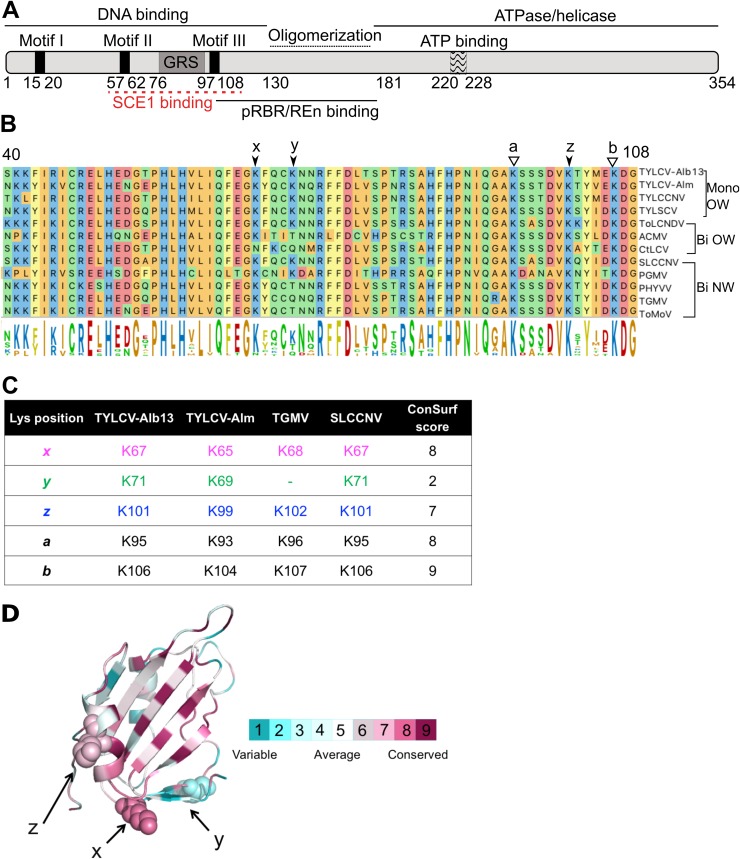
FIG 1.
Lysine residues involved in SCE1 binding are conserved in Rep proteins from different geminiviruses. (A) Diagram of REP with its known functional domains; the red line indicates the region of RepTGMV required for SCE1 binding. (B) Protein sequence alignment of the Reps from different begomoviruses, depicting the region corresponding to residues 40 to 108 in RepTYLCV. The full virus names are indicated in Materials and Methods. The arrows and letters indicate lysines important for RepTGMV to interact with SCE1 and the additional Lys (y) found in some Rep proteins in this domain. Black arrowheads point to residues studied here, and white arrowheads point to Lys residues required for viral replication and not targeted for mutagenesis in this work. Mono and Bi, monopartite and bipartite begomoviruses, respectively; OW and NW, origin of the analyzed viruses (Old World and New World, respectively). (C) Table indicating the corresponding positions and ConSurf scores (see below) of the analyzed Lys residues in the Reps studied here. To each of the Lys residues studied here, a unique color that is also used in the following figures is assigned. (D) Ribbon diagram of the three-dimensional structure model of the N-terminal half of RepTYLCV colored according to the degree of sequence conservation, using a scale from maroon (highest) to cyan (lowest), based on the ConSurf program (97) using an MSA with 337 Rep protein sequences from different geminiviral species and isolates (67).