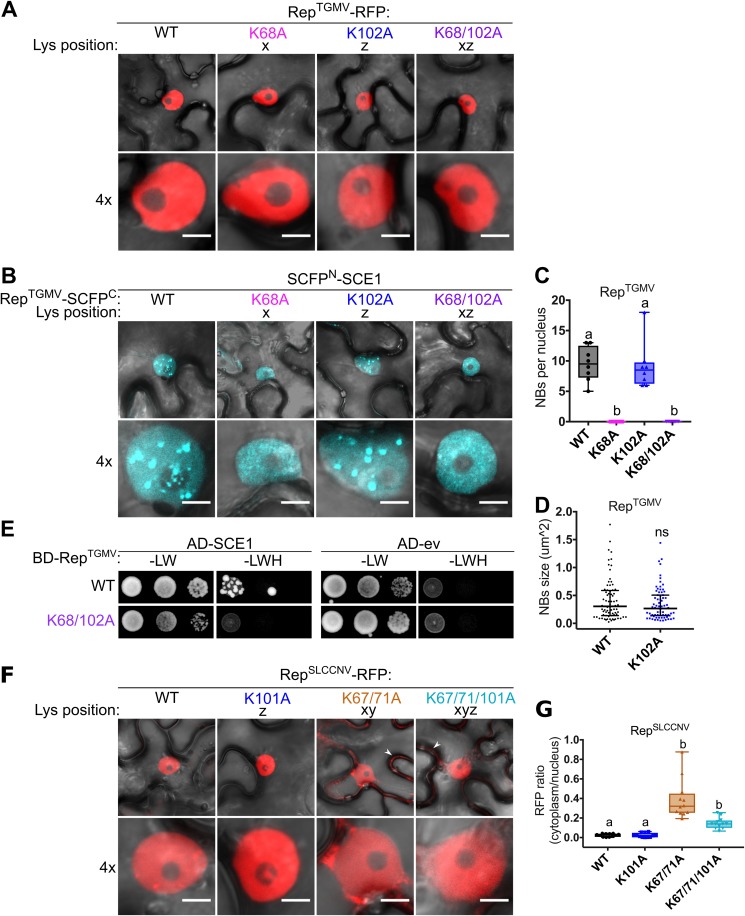
FIG 4.
The lysine residues are not essential for nuclear localization of RepTGMV, while they are in part for RepSLCCNV. (A) RepTGMV-RFP variants reside exclusively in the nucleus, meaning that introduction of K-to-A mutations did not impede the nuclear localization. Scale bars represent 5 μm. (B) BiFC assay showing that WT RepTGMV and K102A interact with SCE1 inside NBs, while K68A and K68/102A mutants yield a diffuse fluorescence signal in the nucleus. (C) Box plot depicting the number of NBs per nucleus in cells expressing the indicated RepTGMV variants (x axis) as a BiFC pair with SCE1. Eight cells were analyzed per sample. (D) As for panel C except that the scatter plot represents the size of NBs. (E) Yeast two-hybrid assay between BDGAL4 fused to RepTGMV WT and K68/102A variants and ADGAL4-SCE1 (left) or ADGAL4-empty vector (ev) (right). –LW, control plate for yeast growth; –LWH, selection plate with medium to test for the interaction. (F) Subcellular localization of RFP-tagged RepSLCCNV WT and K-to-A variants in N. benthamiana upon transient expression using Agrobacterium. White arrowheads indicate cytoplasmic localization of Rep. The scale bars are 5 μm. (G) Box plot depicting the ratio of the RFP fluorescence intensity in the cytoplasm versus nucleus for the images shown in panel F; 12 cells were analyzed per sample. Conditions were similar to those for Fig. 2.