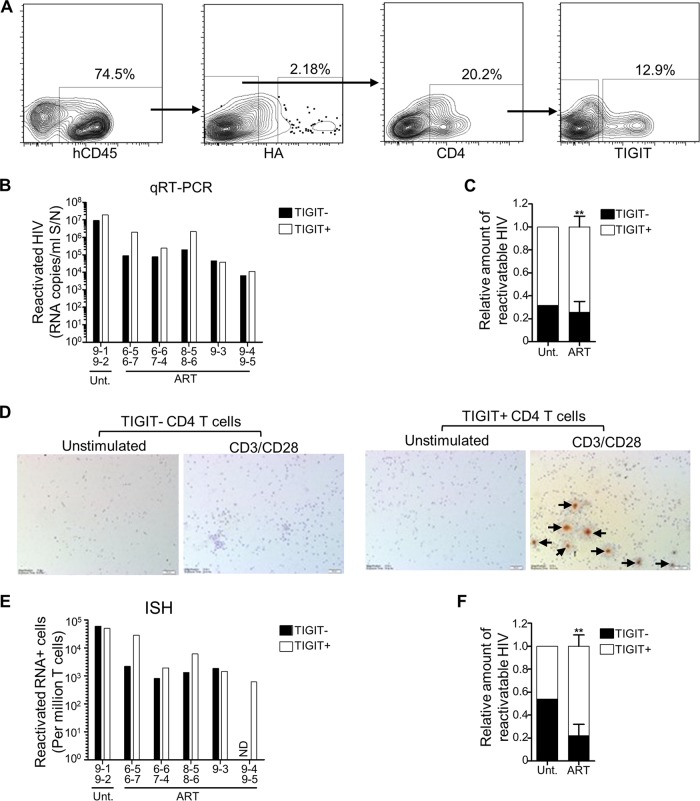
FIG 6.
Latent and reactivatable HIV-1 is enriched in TIGIT+ CD4 T cells. Humanized mice generated from four different cohorts (cohorts 6 to 9) were infected with NL4-3-HA and then either left untreated (n = 2) or switched to oral ART (n = 9). Mouse IDs are denoted on the x axes of relevant graphs. After necropsy, HA– CD45+ CD4+ TIGIT+ and HA– CD45+ CD4+ TIGIT– subsets of spleen cells were sorted by FACS, and equal numbers were plated and cultured in either unstimulating or CD3/CD28 stimulating conditions for the ex vivo latency assay. (A) Representative FACS plots showing the HA– CD45+ CD4+ TIGIT+ and HA– CD45+ CD4+ TIGIT– T cell sorting. (B) Reactivated HIV-1 RNA in culture supernatants for TIGIT– and TIGIT+ subsets from the indicated mice. Due to low recovery, some samples from individual mice were pooled, as indicated. Reactivated HIV values were calculated by subtracting the values from unstimulated cultures from the values of the matched stimulated cultures. (C) Distribution of reactivatable HIV-1 from the qRT-PCR analysis between TIGIT+ and TIGIT– subsets for samples from untreated mice (n = 1) and ART-treated mice (means ± the SEM; **, P < 0.01). (D) Cell pellets from ex vivo latency assay cultures were analyzed for HIV-1 RNA by ISH. Representative images from one sample (combined cells from mouse IDs 6-5 and 6-7) are shown, with arrows indicating HIV-1 RNA+ cells. (E) HIV-1 RNA+ cells from each ISH sample were counted and converted to HIV+ cells/million. Due to low recovery, some cells from individual mice were pooled, as indicated. Values were calculated by subtracting the values from unstimulated cultures from the values of the matched stimulated cultures. ND, not detected. (F) Distribution of reactivatable HIV-1 from the ISH analysis between TIGIT+ and TIGIT– subsets of samples from untreated (n = 1) and ART-treated (n = 5) mice (mean ± the SEM; **, P < 0.01).