The transformation from natural fermentation to synthetic fermentation is essential in constructing a constant food fermentation process, which is the premise for stably making high-quality food. According to flavor-producing and cooccurring functions in dominant microbes, we provided a system-level approach to identify the core microbiota in Chinese light-aroma-type liquor fermentation. In addition, we successfully constructed a synthetic core microbiota to simulate the microbial community succession and flavor compound production in the in vitro system. The constructed synthetic core microbiota could not only facilitate a mechanistic understanding of the structure and function of the microbiota but also be beneficial for constructing a tractable and reproducible food fermentation process.
KEYWORDS: Chinese liquor, cooccurring network, core microbiota, environmental factors, flavor compounds, food fermentation
ABSTRACT
Natural microbiota plays an essential role in flavor compounds used in traditional food fermentation; however, the fluctuation in natural microbiota results in inconsistency in food quality. Thus, it is critical to reveal the core microbiota for flavor compound production and to construct a synthetic core microbiota for use in constant food fermentation. Here, we reveal the core microbiota based on their flavor production and cooccurrence performance, using Chinese light-aroma-type liquor as a model system. Five genera, Lactobacillus, Saccharomyces, Pichia, Geotrichum, and Candida, were identified to be the core microbiota. The synthetic core microbiota of these five genera presented a reproducible dynamic profile similar to that in the natural microbiota. A Monte Carlo test showed that the effects of five environmental factors (lactic acid, ethanol, and acetic acid contents, moisture, and pH) on the synthetic microbiota distribution were highly significant (P < 0.01), similar to those effects on a natural fermentation system. In addition, 77.27% of the flavor compounds produced by the synthetic core microbiota showed a similar dynamic profile (ρ > 0) with that in the natural liquor fermentation process, and the flavor profile presented a similar composition. It indicated that the synthetic core microbiota is efficient for reproducible flavor metabolism. This work established a method for identifying core microbiota and constructing a synthetic microbiota for reproducible flavor compounds. This work is of great significance for the tractable and constant production of various fermented foods.
IMPORTANCE The transformation from natural fermentation to synthetic fermentation is essential in constructing a constant food fermentation process, which is the premise for stably making high-quality food. According to flavor-producing and cooccurring functions in dominant microbes, we provided a system-level approach to identify the core microbiota in Chinese light-aroma-type liquor fermentation. In addition, we successfully constructed a synthetic core microbiota to simulate the microbial community succession and flavor compound production in the in vitro system. The constructed synthetic core microbiota could not only facilitate a mechanistic understanding of the structure and function of the microbiota but also be beneficial for constructing a tractable and reproducible food fermentation process.
INTRODUCTION
Traditional fermented foods are usually produced by natural fermentation containing a multispecies community (1–4). At present, the transformation from natural fermentation to tractable fermentation with the synthetic core microbiota is essential for consistent quality of fermented foods, because only limited genera of microbes in natural microbiota can drive the fermentation process. They not only generate flavor compounds but also maintain microbe interactions which serve to achieve successful food fermentation (5, 6). Thus, revealing the composition of these microbes, which is the core microbiota, is essential for constructing a synthetic microbiota in food fermentation (7).
A series of studies were carried out to identify the core microbiota involved during food fermentation (7–10). Dominant genera were considered to be an essential component in food fermentation (11, 12). For example, a total of 17 genera were identified to be dominant microbes due to their relative abundances in cheese (11). However, dominant genera may not have the ability to produce flavor compounds in food fermentation (13, 14). Researchers have suggested that the identification of core microbiota should also consider microbial flavor compound productivity (7, 15). For example, seven genera were determined to be in the functional core microbiota for the production of flavor compounds in Chinese vinegar fermentation (7).
Recently, we found that the dominant microbes and flavor-producing microbes did not show efficient flavor compound productivity when they were in a mixed culture (13). In contrast, some other microbes were not flavor compound producers, but they showed activity that cooccurred with those flavor-producing microbes, hence leading to an improvement in flavor compounds (13). For example, Pichia membranaefaciens and Bacillus amyloliquefaciens were not efficient flavor compound producers, but they alleviated the competition among flavor compound producers (Saccharomyces cerevisiae, Issatchenkia orientalis, and Bacillus licheniformis) and altered the growth of producers and the production of flavor compounds (13). Moreover, the interaction between microbes plays a vital role in some flavor-producing metabolisms, such as 3-(methylthio)-1-propanol and dimethyl disulfide (16). As a consequence, we suggest that besides flavor compound productivity, microbial interactions should also be considered in identifying the core microbiota. Moreover, microbial interaction is a critical factor for maintaining the cooccurring in microbial communities, and cooccurring network analysis is an effective tool for studying the microbial interaction (17, 18).
Thus, to overcome the problem of inaccurate definition of core microbiota in fermented foods, we developed a comprehensive method to identify the core microbiota in natural food fermentation that combined flavor production and cooccurring network analyses. We examined the activity of the core microbiota, including their interaction with environmental factors, and flavor compound production. Light-aroma-type liquor, generated by a natural fermentation process, is a favorite alcoholic beverage in China (19). In this work, using Chinese light-aroma-type liquor fermentation as a model system, we provided a strategy to identify the core microbiota and constructed a synthetic microbiota using the core microbiota. Because Chinese light-aroma-type liquor, a typical and popular fermented food, is made from spontaneous fermentation involving multiple microbes and complex interactions between microbes (12, 20), this type of fermentation can produce unique food flavor and taste characteristics (1). Also, it is also one of the three typical type liquors in China (sauce-aroma-, strong-aroma-, and light-aroma-type liquors). In addition, a smaller brewing container can be used, it requires a shorter fermentation time, and it is easy to observe. For example, the fermentation container volume and fermentation time of sauce-aroma-type liquor are about 22.3 m3 and 240 days, but the volume and fermentation time of light-aroma-type liquor are only 0.46 m3 and 60 days. Therefore, it is beneficial to take Chinese liquor production as a model system and establish a method to define the core microbiota to construct a synthetic microbiota to elucidate the metabolism of fermented foods.
RESULTS
Microbial diversity during the fermentation process.
Across all samples, altogether, 453,217 and 677,563 high-quality sequences were identified for bacteria and fungi after quality control. Meanwhile, a total of 722 and 1,504 operational taxonomic units (OTUs) were obtained for bacteria and fungi with 97% similarity. A total of 49 bacterial genera and 34 fungal genera were identified in the fermentation process (see Data Set S1 in the supplemental material). All Good’s coverage values of the samples were over 99.80% (see Table S1 in the supplemental material), which indicated that the sequences represented the majority of the microbiota in the fermentation process (21). The average bacterial α-diversity (Chao1 richness and Shannon diversity) declined along with fermentation time on the whole, but there was a fluctuation on day 15 (Table S1). On the contrary, the average fungal α-diversity (Chao1 richness and Shannon diversity) increased along with fermentation time on the whole, but there was a fluctuation on day 5 (Table S1).
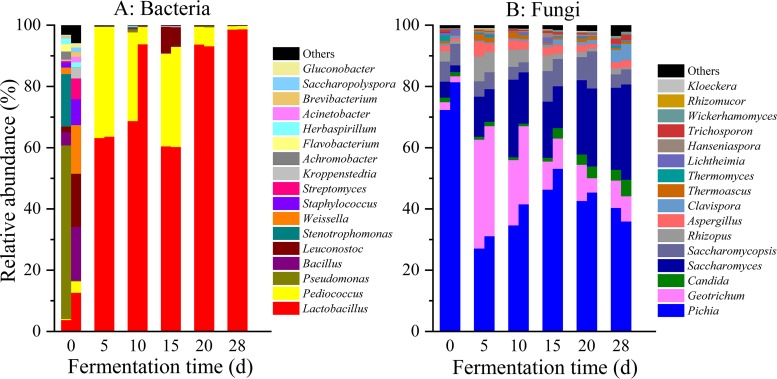
As for bacteria (Fig. 1A), at the early stage of fermentation (day 0), Pseudomonas and Bacillus were the predominant genera (average abundances, ≥10%) (22), whereas Lactobacillus, Pediococcus, Leuconostoc, Weissella, Stenotrophomonas, Staphylococcus, Streptomyces, Kroppenstedtia, Herbaspirillum, Achromobacter, Flavobacterium, and Brevibacterium were the subdominant genera (1% ≤ average abundance < 10%). During the middle stage of fermentation (days 5 to 15), Lactobacillus and Pediococcus became the predominant genera, and Leuconostoc was the subdominant genus at day 15. At the late stage of fermentation (days 20 to 28), only Lactobacillus was the predominant genus, and Pediococcus became the subdominant genus. As for fungi (Fig. 1B), Pichia was the predominant genus (average abundance, ≥10%) during the whole fermentation process. Geotrichum (days 5 to 10) and Saccharomyces (day 10) were the predominant genera, and Saccharomycopsis, Rhizopus, Clavispora, Candida, Aspergillus, Thermomyces, Thermoascus, Trichosporon, and Lichtheimia were the subdominant genera (1% ≤ average abundance < 10%) at different stages of fermentation.
FIG 1.
Distribution of the relative abundances of bacterial (A) and fungal (B) genera during the fermentation in the in situ system. Only those genera that had an average abundance greater than 1% are indicated. Genera with less than 1% abundance are combined and shown in “others” category. d, days.
Through statistical analysis of all communities sampled, only 17 bacterial and 16 fungal genera were found at greater than 1% average abundance, as these were defined as the dominant microbiota (9). A ubiquitously distributed microbiota is usually defined as being present in most samples (9, 23). Therefore, we defined microbes which exist in more than 50% of samples within a total of 14 samples as ubiquitously distributed microbiota (9). Two genera of bacteria (Lactobacillus and Pediococcus) and eight genera of fungi (Pichia, Geotrichum, Saccharomyces, Saccharomycopsis, Rhizopus, Aspergillus, Candida, and Thermoascus) were identified to be ubiquitously distributed dominant microbiota (Table S2).
Identification of core microbiota.
Flavor compounds are very important indicators of liquor quality (24, 25). A total of 41 kinds of flavor compounds were identified during the fermentation process (Fig. 2), including four alcohols, two carbonyl compounds, five acids, 20 esters, nine aromatic compounds, and one heterocyclic compound.
FIG 2.
Heatmap of flavor metabolites and hierarchical clustering in the in situ fermentation process. Flavor compounds were transformed by z-score. Clustering analysis was performed using the Pearson correlation coefficient and Euclidean distance based on the flavor contents during the fermentation process.
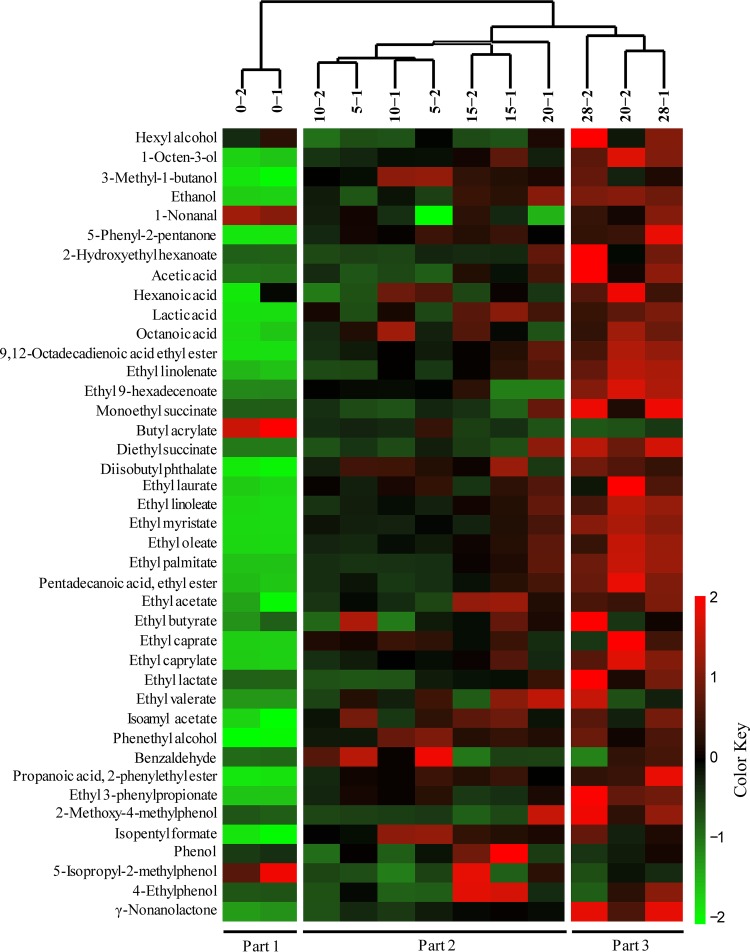
The concentrations of flavor compounds were converted into a heatmap, and hierarchical cluster analysis was performed. As shown in Fig. 2, the hierarchical clustering results showed that the fermentation process consisted of three parts based on the dynamic profile of flavor compounds, part 1 (day 0), part 2 (days 5 to 20), and part 3 (days 20 to 28).
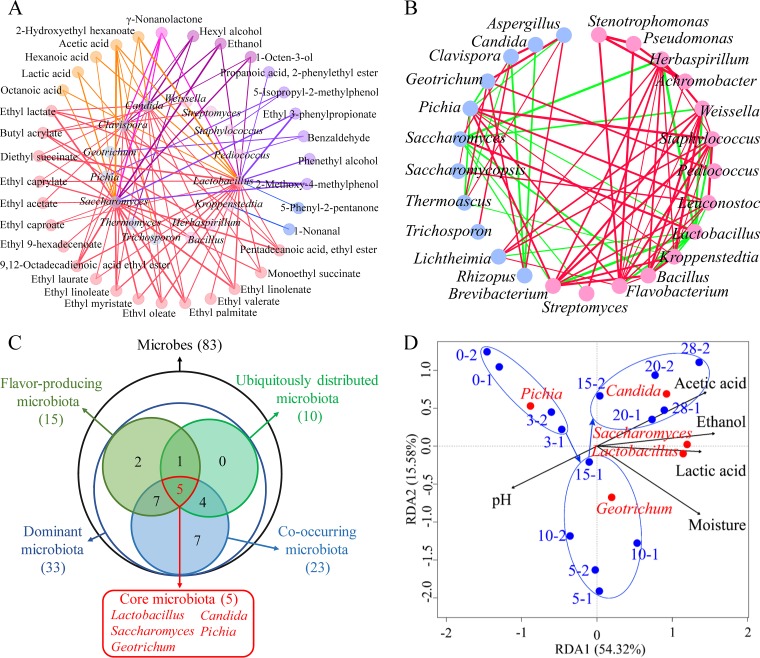
Most flavor compounds interact with microbes in food fermentation. Network correlation analysis is a powerful tool to investigate the potential interactions between microbes and flavor compounds (26). Thus, we calculated the Spearman correlation coefficient between 33 dominant genera and 41 flavor compounds and chose a coefficient (ρ) of >0.5 and significance (P value) of <0.05 (27, 28) as demonstrating strongly correlated nodes of the network (Fig. 3A). Eight bacterial and seven fungal genera were significantly correlated (P < 0.05, ρ > 0.5) with 34 kinds of flavor compounds, indicating that these 15 genera are the flavor-producing microbiota (Table S3). Among them, Lactobacillus, Saccharomyces, Clavispora, and Candida were significantly correlated (P < 0.05, ρ > 0.5), with 26, 26, 16, and 14 kinds of flavor compounds, respectively (Fig. 3A).
FIG 3.
Identification of the core microbiota in the in situ system. (A) Correlation network between microbial genera and flavor compounds during the fermentation process in the in situ system. Inner circle nodes represent microbes (light-red nodes represent bacteria genera, and light-blue nodes represent the fungi genera), and outer circle nodes represent flavor compounds (different colors represent different flavor types). The thickness of the lines is proportional to the value of Spearman’s correlation (ρ > 0.5, P < 0.05). The colors of the lines are the same as the flavor nodes. (B) Correlation network of cooccurring genera in dominant microbiota. Statistical significance (P < 0.05) and Spearman correlation coefficient (|ρ| > 0.5) indicate the correlations. Light-red nodes represent bacterial genera, and light-blue nodes represent fungal genera. Green and red lines indicate negative and positive interactions, respectively, between genera. The thickness of the lines represents the strength of interaction. (C) Venn diagram of the core microbiota. Different circles represent different genus categories. (D) RDA of fermentation process. Blue dots represent the times of fermentation. Red dots represent the core microbiota. Black arrows point to the different environmental factors. Percentages on the axes represent the eigenvalues of principal components.
Cooccurrence network analysis allows identification of the cooccurring microbiota (17). We calculated the Spearman correlation coefficients of 33 dominant genera. A Spearman’s correlation coefficient (|ρ|) of >0.5 and P value of <0.05 were considered to represent a valid cooccurrence event (17, 18, 26, 29, 30). Through the cooccurrence network analysis, a total of 25 nodes and 149 edges were obtained (|ρ| > 0.5, P < 0.05), and the average network clustering coefficient was 0.696, which suggested that the network had nodular structures. In Fig. 3B, different genera are divided into different nodular structures. A total of 23 genera demonstrated a high degree of connection (≥4 edges per node) (26) and were defined as the cooccurring microbiota, including Flavobacterium, Lactobacillus, Brevibacterium, Herbaspirillum, Pichia, Staphylococcus, Bacillus, Weissella, Kroppenstedtia, Leuconostoc, Saccharomyces, Aspergillus, Clavispora, Geotrichum, Lichtheimia, Thermoascus, Rhizopus, Achromobacter, Pseudomonas, Stenotrophomonas, Candida, Saccharomycopsis, and Streptomyces (Table S4). In the cooccurrence network, Lactobacillus and Saccharomyces spp. were mainly negatively correlated (ρ < −0.5) with other microbes (except Clavispora), but they showed a positive correlation with each other.
In summary, we obtained ubiquitously distributed dominant microbiota (10 genera), flavor-producing microbiota (15 genera), and cooccurring microbiota (23 genera). Five genera, Lactobacillus, Saccharomyces, Geotrichum, Candida, and Pichia, existed in all three different microbiota populations (Fig. 3C). Due to their high relative abundances and frequencies, their contributions to flavor production, and the stable microbial network, they were defined as the core microbiota in liquor fermentation.
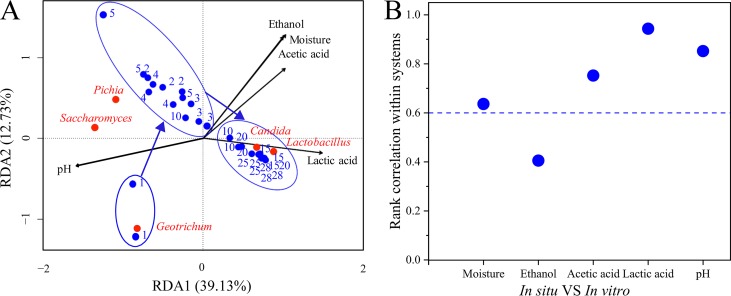
The impact of five environmental factors on the core microbiota was analyzed, including lactic acid content, ethanol content, acetic acid content, moisture, and pH (Table S5). Variation partitioning analysis (31) was used to calculate the contributions of these environmental factors. The results showed that these five environmental factors accounted for 87.18% of core microbiota variation in the in situ systems (Table S6). Partial redundancy analysis (RDA) was used to identify the effects of these factors on the core microbiota (Fig. 3D). Acetic acid content, ethanol content, and lactic acid content were positively correlated with Lactobacillus, Saccharomyces, and Candida at the end of fermentation. A Monte Carlo replacement test (Table S7) verified the result that these factors were significantly correlated with the core microbiota (P < 0.05). It indicated that these five environmental factors had a significant influence on the core microbiota.
Reproducible dynamic profile of microbiota in synthetic core microbiota.
In this study, we provided a system-level approach to identify the core microbiota in Chinese light-aroma-type liquor fermentation and obtained five different core genera during the whole fermentation stage, Lactobacillus, Pichia, Geotrichum, Candida, and Saccharomyces. Due to the diversity of genera, it was considered feasible that isolated species represented certain taxa. For example, cheese rind isolates that represented the most abundant taxa were applied to construct in vitro communities of cheese rind (9). Using 16S rRNA and internal transcribed spacer (ITS) amplification sequence data, when the sequence identity was greater than 99% compared to the type and reference strains, assignment to the species level was performed (32). Thus, we identified one species with the highest relative abundance in each corresponding genus (Fig. S1 and Table S8) and used that as the starter species of the synthetic microbiota, including Lactobacillus acetotolerans, Pichia kudriavzevii, Geotrichum candidum, Candida vini, and Saccharomyces cerevisiae. Lactobacillus acetotolerans is a functional microorganism in the fermentation of different kinds of liquors (strong-aroma-type liquor, light-aroma-type liquor, and Japanese sake) (32–34). For example, Lactobacillus acetotolerans appeared to play a key role during the Chinese strong-aroma-type liquor fermentation (32), and it had positive relationships with most chemical components that contribute to the quality and flavor of liquor (35). Pichia kudriavzevii contributes to the functionality (acids and esters) of foods during fermentation, and it can improve the sensory and some functional properties of the cereal-based substrate during fermentation (36). Geotrichum candidum can produce lipases which would be important for the production of fruity-aroma compounds (37). Candida vini had been shown to contribute to fatty acid production (38). Saccharomyces cerevisiae is an important strain of ethanol fermentation in Chinese liquor fermentation (39). Therefore, we chose the above-mentioned five species for the synthetic microbiota experiment.
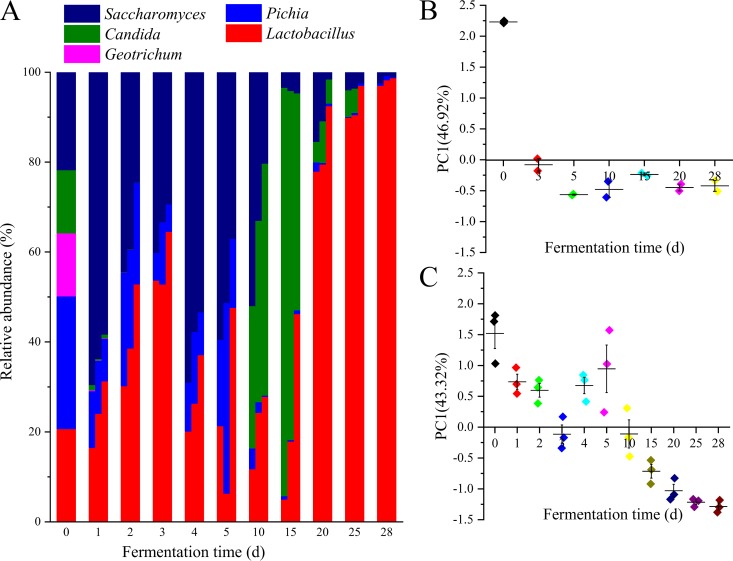
We inoculated approximately equal numbers of each species in the five core genera together into fermented grains in the in vitro system (Fig. 4A). Lactobacillus became the predominant bacterial genus in the in vitro system as fermentation proceeded (Fig. 4A and S2B), which was similar to the makeup in the in situ system (Fig. S2A and S3). Saccharomyces and Pichia were the dominant genera early in (1 to 5 days) and at the end of (28 days) the fermentation process, which was similar to the makeup in the in situ system (Fig. 4A and S2C and D). Candida was the dominant fungal genus in the middle of the fermentation process (10 to 25 days). It revealed that the successive direction of the in vitro system (Fig. 4B) in the principal component is consistent with that of the in situ system over a 28-day fermentation period (Fig. 4C), which demonstrated a highly reproducible microbial succession pattern in in vitro liquor fermentation.
FIG 4.
Reproducible dynamic profile of microbiota in synthetic core microbiota. (A) Distribution of the abundance of genera during the fermentation in the in vitro system. (B) In situ, the change of principal component in time gradient. (C) In vitro, the change of principal component in time gradient.
The impact of the environmental factors on the synthetic microbiota was also analyzed (Table S9). The results from variation partitioning analysis of the five environmental factors (lactic acid content, ethanol content, acetic acid content, moisture, and pH) could be explained in 53.65% of cases in the in vitro system (Table S6). This percentage showed that these five factors drove the variation of the synthetic core microbiota. RDA showed that pH was negatively correlated with the other environmental factors, which was the same in the in situ system (Fig. 5A). Lactic acid content, acetic acid content, ethanol content, and moisture were positively correlated with each other, which is consistent with the in situ system. A Monte Carlo test also showed that the interpretation of these five environmental factors on the synthetic microbiota distribution was that they were highly significant (P < 0.01) (Table S7). Through the change in correlation analysis regarding environmental factors in the two systems with temporal dynamics (Fig. 5B), we found that five environmental factors had a positive correlation (ρ > 0) with the core microbiota; especially, moisture, acetic acid content, lactic acid content, and pH had a strong correlation (ρ > 0.6) between the in situ and in vitro systems. These results indicated that the effects of environmental factors on the core microbiota were similar in the in vitro and in situ systems.
FIG 5.
RDA of fermentation process in the in vitro system and the relationship of environmental factors within the in situ system. (A) RDA of fermentation process in the in vitro system (as described for Fig. 3D). (B) Similarity of the in situ and in vitro systems. The y axis represents the Spearman correlation coefficient between the corresponding environmental factors in the two systems. The x axis represents the environmental factors.
Reproducible flavor metabolism in synthetic core microbiota.
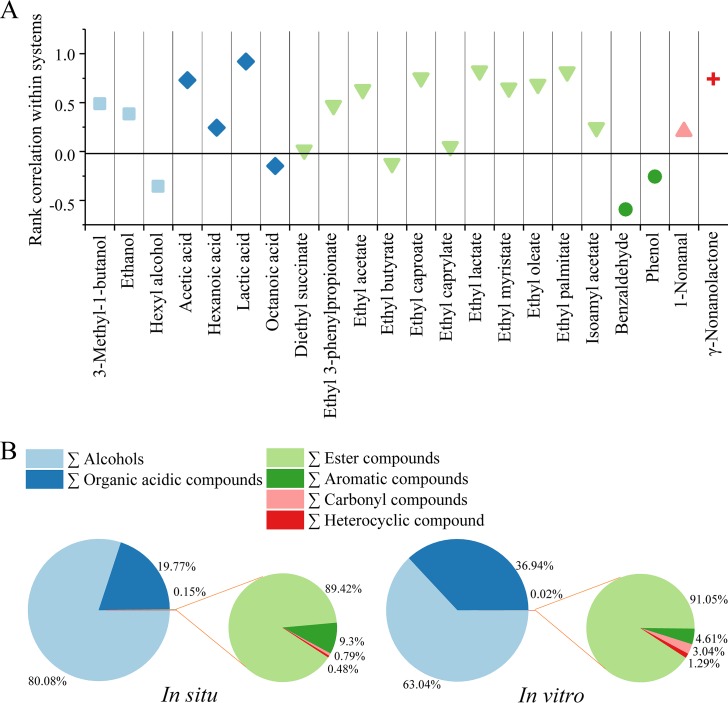
The flavor compound production in the synthetic microbiota was determined, and 22 flavor compounds were identified in the in situ system (Fig. 6A). The in vitro generation of flavor compounds can be divided into three parts (Fig. S4), part 1 (days 0 to 3), part 2 (days 4 to 10), and part 3 (days 15 to 28). The temporal dynamics was similar to that in the in situ system.
FIG 6.
Reproducible flavor metabolism in synthetic core microbiota. (A) Similarity of the two systems in 22 kinds of alcohols, acids, and esters. The y axis represents the Spearman correlation coefficient (ρ) of the flavor generation along the time axis in the two systems. The x axis represents the flavors in two systems. (B) Proportions of six kinds of flavor compounds in the in situ and in vitro systems.
The Spearman correlation coefficient (ρ) of the 22 flavor compound generation in the two systems was calculated in the fermentation. The result showed that 17 kinds of flavor compounds (proportion = 77.27%) had a positive correlation (ρ > 0) with generation on the temporal dynamics in the two systems (Fig. 6A). The different flavor classifications had similar proportions in the two systems (Fig. 6B), in which the proportions of alcohols and acids accounted for more than 99.85% in the total flavor compounds. This indicated that the flavor metabolism could be reproduced in the in vitro system using the synthetic core microbiota.
DISCUSSION
The core microbiota present in food fermentation is of great importance to knowledge of the quality and characteristics of foods. Many molecular and ecological approaches have been used to characterize the core microbiota (22, 40–42). In this work, we chose microbial communities in the Chinese light-aroma-type liquor fermentation process as a model system and provided a system-level method for identifying the core microbiota in natural food fermentation. This is a prudent way to examine the characteristics of the dynamic success of the microbiota, the effect of the environmental factors, and the profile of flavor compound production. Among these compounds, we did not detect the detrimental flavors in Chinese light-aroma-type liquor fermentation. Most of these flavors have pleasant aromatic smells, such as those from ethyl acetate (pineapple), ethyl lactate (fruity), 1-octen-3-ol (mushroom), octanoic acid (cheesy), ethyl 3-phenylpropanoate (floral), γ-nonanolactone (coconut), etc. (24, 43). Although some of these flavors are unpleasant flavors, they form a special style of products at low concentrations, such as acetic acid (acidic, vinegar), hexanoic acid (sweaty), ethyl oleate (fatty), 3-methyl-1-butanol (malty), etc. (24, 43). We constructed a reproducible synthetic core microbiota and compared it with the natural microbiota for liquor fermentation, which we hoped would help us establish a tractable food fermentation system.
In the in vitro system, the alcohol (without ethanol) and acid contents were a bit higher than those in the in situ system (Data Sets S2 and S3), whereas ester contents were lower than those in the in situ system (P < 0.001). This may be due to the low concentration of esterification strains in the in vitro system. We also observed slight differences in the microbiota between the in situ and in vitro systems. For example, a succession of Saccharomyces spp. appeared to proceed much more quickly (Fig. S2C and D), and Candida spp. showed a higher relative abundance in the later fermentation in the in vitro system (Fig. 4 and S2D). The difference might result from a higher initial proportion of these genera in the in vitro system. Therefore, the initial compositions of the core microbiota should be optimized in synthetic core microbiota fermentation. Different species and different strains of microorganisms belonging to the same genus may have different metabolic functions. Therefore, more functional strains should be isolated. However, the same strain in single fermentation and mixed fermentation may show completely different metabolic patterns (16). Therefore, the target functional strains should be synthetically optimized by extensive statistical analysis.
Besides the liquor fermentation system, the methods for identifying the core microbiota and constructing a synthetic microbiota for food fermentation can also be used in a variety of food fermentation processes. Various food fermentations share core microbiota members because these members present similar functions. For example, Lactobacillus spp. were confirmed to be the core microbe in fermentations of vinegar, liqueur, cheese, pickle, and so on (44–46). They contributed amino acids (glutamic acid, alanine, valine, etc.), organic acids (acetic acid, lactic acid, etc.), and other flavor compounds (7, 47–49). They also interacted with other microbes, such as species of Bacillus, Aspergillus, and Luteococcus, hence regulating their flavor compound production (46, 49–51). Pichia spp. are widely used in food fermentation, such as for wine and beer (52, 53; U.S. patent application 20160010042). They are considered to be essential producers of esters (55). Pichia spp. can also maintain the cooccurrence of the community (13), which was similar to that shown in Fig. 3B. Geotrichum spp. can produce lipases, which are important for the production of fruity-aroma compounds, such as ethyl esters of acetic acid, propionic acid, butyric acid, and isobutyric acid (56, 57). Saccharomyces spp., as ethanol producers, are widely used in the production of liquor and other alcoholic beverages (58). They drove the development direction of the microbiota, together with Lactobacillus spp. (acid producers) (59, 60). Candida spp. were widely used in food fermentation due to their production of various lipases (Antarctica lipase A, rugosa lipases, glucose ester synthesis lipase, etc.) (61–63). When Candida and Saccharomyces spp. were cocultured in wine fermentation, they produced greater amounts of esters and glycerol than with single Saccharomyces culture (64). These studies indicated that most of the microbes in the core microbiota had similar functions in different food fermentations.
The transformation from natural fermentation to synthetic fermentation is essential to construct a tractable food fermentation process, which is the premise for stably making high-quality foods. We provided a system-level approach to identify the core microbiota in food fermentation and constructed a synthetic microbiota for reproducible flavor metabolism. The synthetic microbiota was hoped to provide a chance for us to define the mechanisms underlying the microbial interaction and contribution to flavor compounds in the food microbiota. It is also important to manipulate the synthetic microbiota and then control the quality of fermented foods.
MATERIALS AND METHODS
Sample collection.
Samples were collected from a local liquor distillery (Shanxi Xinghuacun Fenjiu Distillery Co. Ltd., Shanxi, China). For liquor fermentation, the steamed grains were mixed with starter at a ratio of 9:1 (wt/wt) and put into earthenware jars. Then, the jars were sealed for 28 days of fermentation. For the survey of microbial diversity, a total of 12 samples (100 g each sample) were collected from 2 jars in the center of the layer (0.5 m deep) at different fermentation times (days 0, 5, 10, 15, 20, and 28) in April 2016. When we took the samples, we opened a small part of the space and immediately filled the sampling space again after taking the samples. In the next sampling, we changed to a different sampling site to avoid possible interference between the different samples. All samples were stored at −20°C for further DNA extraction and physicochemical parameter determination.
DNA extraction, qualification, and sequencing analysis.
Each sample (5.00 g) was used to extract genomic DNA using the E.Z.N.A. soil DNA kit (Omega Bio-tek, Norcross, GA), according to the manufacturer’s instruction. The V3-V4 region of the 16S rRNA bacterial gene was amplified using the universal primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GACTACHVGGGTWTCTAAT-3′) (65). For fungi, the ITS2 region was amplified using the primers ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′) and ITS3 (5′-GCATCGATGAAGAACGCAGC-3′) (66). These primers added an 8-nucleotide barcode sequence unique to each sample. The PCR mixtures contained 2.5 μl of 10 × Pyrobest buffer, 2 μl of 2.5 mM dinucleoside triphosphates (dNTPs), 1 μl of each primer (10 μM), 0.4 U of Pyrobest DNA polymerase (TaKaRa Holdings, Inc., Shiga, Japan), 15 ng of template DNA, and double-distilled water (ddH2O) up to a volume of 25 μl. Amplification was performed using a previously described method (42, 67). Then, amplicons were pooled in equimolar quantities and subjected to high-throughput sequencing using a MiSeq benchtop sequencer for 2 × 300-bp paired-end sequencing (Illumina, San Diego, CA). The EzBioCloud and Central Bureau of Fungal Cultures (CBS-KNAW) databases were used for sequence alignment of bacteria and fungi.
Population determination by real-time quantitative PCR.
The populations of yeast and lactic acid bacteria (LAB) in liquor fermentation were determined by real-time quantitative PCR (qPCR). Genomic DNA of samples were used as the template. For yeast, the sequences were amplified using YEASTF (5′-GAGTCGAGTTGTTTGGGAATGC-3′) and YEASTR (5′-TCTCTTTTCCAAAGTTCTTTTCATCTTT-3′) primers (68). For LAB, the sequences were amplified using Lac1 (5′-AGCAGTAGGGAATCTTCCA-3′) and Lac2 (5′-ATTYCACCGCTACACATG-3′) primers (69). qPCR was performed using the StepOnePlus instrument (Applied Biosystems, CA, USA) (16).
Sequence processing.
All the raw MiSeq-generated sequences were processed via QIIME (version 1.8) (70). Briefly, high-quality sequences were made by removing sequences with >2 ambiguous bases, >10 homopolymers, primer mismatches, average quality scores of <20, and lengths (excluding the primer or barcode region) of <50 bp. Chimeras were removed using USEARCH (version 10) (71). The trimmed sequences were clustered into operational taxonomic units (OTUs) with 97% sequence (72), and the Shannon index and Chao1 estimator value were calculated using UCLUST (version 1.2.22) (73, 74).
Analysis of environmental factors and flavor compounds.
Moisture was measured by determining weight loss by drying 10 g of each sample at 105°C for 3 h (sufficient to ensure constant weight) (75). The pH was measured at a 1:2.5 (wt/vol) ratio in double-distilled water (ddH2O) with the laboratory pH meter-FE20 (Mettler Toledo, Shanghai, China) (75). Five-gram samples were added to 10 ml ddH2O, put in an ultrasonic cleaner (AS30600B; Autoscience, Tianjin, China) for 30 min, and then centrifuged at 8,000 × g for 10 min. After filtering using a 0.2-μm-pore-size filter, the filtrate was used to analyze the concentrations of flavor compounds and acids. The flavor compound content was detected using gas chromatography-mass spectrometry (6890N GC system and 5975 mass-selective detector; Agilent, Santa Clara, CA) (42). The ethanol content was determined by high-performance liquid chromatography (HPLC; Agilent 1200) using an Aminex HPX-87H column (Bio-Rad, Hercules, CA) (76). The contents of lactic acid and acetic acid were measured using reversed-phase ultraperformance liquid chromatography (UPLC; H-class system; Waters, Milford, MA) with chromatographic Atlantis T3 (4.6 mm by 150 mm, 3 μm) column (Waters) and RP-C18 SecurityGuard column (4.0 mm by 3.0 mm; Phenomenex, Inc., Torrance, CA). The UV detection wavelength was 210 nm. The column temperature was 30°C. The injection volume was 10 μl. The mobile phase was 10 mmol/liter NaH2PO4 (pH 2.7), and the flow velocity was 0.8 ml/min.
Strains.
The predominant microbes isolated from the liquor fermentation process, Lactobacillus acetotolerans, Pichia kudriavzevii, and Candida vini, were deposited in the China General Microbiological Culture Collection Center (CGMCC) with strain names 14086, 12418, and 2.2018, respectively. Saccharomyces cerevisiae was deposited in the China Center for Type Culture Collection with the strain name CCTCC M2014463. Geotrichum candidum is a laboratory strain, XY7.
Liquid fermentation.
Sorghum extract was used as seed fermentation broth (40). The extract was diluted with distilled water to give a sugar concentration of about 90 g/liter and then autoclaved at 115°C for 15 min. One hundred milliliters of medium was added to 150-ml conical flasks, inoculated with a loop of the target strain, and then incubated for 48 h at 30°C (yeast) and 24 h at 37°C (LAB). The microscopy was used to continuously count until 108 CFU/ml seed fermentation broth was obtained.
Solid-state fermentation.
Sorghum (400 g) was added to 500 ml of water in a 3,000-liter beaker, and we mixed the liquefied enzyme (10 U/g) in boiling water (100°C) for 2 h and added glucoamylase (50 U/g); the mixture was maintained for 4 h at 60°C. We then reduced the sugar of the sorghum extracts about 50 times to ∼90 g/kg. The beaker was autoclaved at 115°C for 15 min. After cooling, seed fermentation broth was added to the beaker at a concentration of 1 × 105 CFU/g wet sorghum, and then experiments were carried out in 150-ml conical flasks which contained 100 g of sorghum. The flasks were then sealed and incubated at 30°C. In order not to interrupt the fermentation process, 30 flasks were used for fermentation, under the above-mentioned experimental conditions, and three flasks were randomly selected under the same fermentation conditions at 1, 2, 3, 4, 5, 10, 15, 20, 25, and 28 days. After fermentation, the sorghum samples were used to enumerate different strains, and the rest of the samples were withdrawn and stored at −20°C for analysis of environmental factors and flavor compounds.
Enumeration of different strains.
After fermentation, 10 g sorghum was added to 25 ml phosphate-buffered saline (PBS; 0.01 M [pH 7.2]), followed by vortexing at 3,000 rpm for 30 s (Dragonlab MX-E, Beijing, China) and at 4°C for 30 min. The supernatant was diluted in a gradient and plate spread. Four kinds of yeast enumeration methods were carried out on Wallerstein laboratory nutrient (WLN) medium (77), in which the strains showed different macroscopic features (texture, surface, margin, and color). Lactobacillus enumeration was carried out on de Man-Rogosa-Sharpe (MRS) broth (34). Standard deviations were calculated from triplicate repetitions of the enumerations.
Statistical analysis.
Standard statistical analyses were conducted with XLSTAT (version 19.02.42992). Heatmap development, variation partitioning analysis, redundancy analysis (RDA), and the Monte Carlo permutation test were performed using the R program (version 3.4.0). In the heatmap, flavor compounds were transformed by z-score. Clustering analysis was performed using the Pearson correlation coefficient, and Euclidean distance was based on the flavor compound content during the fermentation process. The variation partitioning analysis identified five environmental factors and the average abundances of five microbes. In constrained ordination, redundancy analysis was used to identify the relationship of samples, environmental factors, and microbes. The Monte Carlo permutation test was used to examine the significance of the correlation between environmental factors and species distribution. All analyses were performed using functions in the vegan package (version 2.4-3) (78). The Spearman correlation coefficient (ρ) and paired-sample t test were calculated with SPSS Statistics 22, in which a ρ value of >0.6 and ρ value of >0.8 were strongly and highly correlated, respectively. Creation of visualizations of flavor compound and microbe interactions and cooccurring analysis were performed with Gephi (version 0.9.1) (22).
Data availability.
The fungal and bacterial raw sequence data were deposited in the DNA Data Bank of Japan (DDBJ) database under the accession numbers DRA005471 and DRA005474, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shanxi Xinghuacun Fenjiu Distillery Co., Ltd., for the samples and Peng Wang and Jianchun Lin for the use of R and sample collection.
This work was supported by the National Natural Science Foundation of China (NSFC) (grant 31530055), National Key R&D Program of China (grants 2018YFD0400402 and 2016YFD0400500), Jiangsu Province Science and Technology Project (grant BE2017705), China Postdoctoral Science Foundation (grant 2017M611702), the Postgraduate Research & Practice Innovation Program of Jiangsu Province (grant KYCX18_1798), National First-Class Discipline Program of Light Industry Technology and Engineering (grant LITE2018-12), the Priority Academic Program Development of Jiangsu Higher Education Institutions, the 111 Project (grant 111-2-06), and the Collaborative Innovation Center of Jiangsu Modern Industrial Fermentation.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03090-18.
REFERENCES
- 1.Smid EJ, Lacroix C. 2013. Microbe-microbe interactions in mixed culture food fermentations. Curr Opin Biotechnol 24:148–154. doi: 10.1016/j.copbio.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Bokulich NA, Thorngate JH, Richardson PM, Mills DA. 2014. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci U S A 111:139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meersman E, Steensels J, Mathawan M, Wittocx PJ, Saels V, Struyf N, Bernaert H, Vrancken G, Verstrepen KJ. 2013. Detailed analysis of the microbial population in Malaysian spontaneous cocoa pulp fermentations reveals a core and variable microbiota. PLoS One 8:e81559. doi: 10.1371/journal.pone.0081559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodríguez-Lerma GK, Gutiérrez-Moreno K, Cárdenas-Manríquez M, Botello-Álvarez E, Jiménez-Islas H, Rico-Martínez R, Navarrete-Bolaños JL. 2011. Microbial ecology studies of spontaneous fermentation: starter culture selection for prickly pear wine production. J Food Sci Technol 76:M346–M352. doi: 10.1111/j.1750-3841.2011.02208.x. [DOI] [PubMed] [Google Scholar]
- 5.Holzapfel W. 2002. Appropriate starter culture technologies for small-scale fermentation in developing countries. Int J Food Microbiol 75:197–212. doi: 10.1016/S0168-1605(01)00707-3. [DOI] [PubMed] [Google Scholar]
- 6.Giraffa G. 2004. Studying the dynamics of microbial populations during food fermentation. FEMS Microbiol Rev 28:251–260. doi: 10.1016/j.femsre.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Wang ZM, Lu ZM, Shi JS, Xu ZH. 2016. Exploring flavour-producing core microbiota in multispecies solid-state fermentation of traditional Chinese vinegar. Sci Rep 6:26818. doi: 10.1038/srep26818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sieuwerts S, de Bok FAM, Hugenholtz J, van Hylckama Vlieg JET. 2008. Unraveling microbial interactions in food fermentations: from classical to genomics approaches. Appl Environ Microbiol 74:4997–5007. doi: 10.1128/AEM.00113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolfe BE, Button JE, Santarelli M, Dutton RJ. 2014. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell 158:422–433. doi: 10.1016/j.cell.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Filippis F, La Storia A, Stellato G, Gatti M, Ercolini D. 2014. A selected core microbiome drives the early stages of three popular Italian cheese manufactures. PLoS One 9:e89680. doi: 10.1371/journal.pone.0089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfe BE, Dutton RJ. 2015. Fermented foods as experimentally tractable microbial ecosystems. Cell 161:49–55. doi: 10.1016/j.cell.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 12.Hu XL, Du H, Ren C, Yan X. 2016. Illuminating anaerobic microbial community and cooccurrence patterns across a quality gradient in Chinese liquor fermentation pit muds. Appl Environ Microbiol 82:2506–2515. doi: 10.1128/AEM.03409-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Q, Ling J, Xu Y. 2014. Starter culture selection for making Chinese sesame-flavored liquor based on microbial metabolic activity in mixed-culture fermentation. Appl Environ Microbiol 80:4450–4459. doi: 10.1128/AEM.00905-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armstrong MS, Boyd CE, Lovell RT. 1986. Environmental factors affecting flavor of channel catfish from production ponds. N Am J Aquac 48:113–119. doi:. [DOI] [Google Scholar]
- 15.Pothakos V, Illeghems K, Laureys D, Spitaels F, Vandamme P, De Vuyst L. 2016. Acetic acid bacteria in fermented food and beverage ecosystems, p 73–99. In Matsushita K, Toyama H, Tonouchi N, Okamoto-Kainuma A (ed), Acetic acid bacteria. Springer, Tokyo, Japan. [Google Scholar]
- 16.Liu J, Wu Q, Wang P, Lin JC, Huang L, Xu Y. 2017. Synergistic effect in core microbiota associated with sulfur metabolism in spontaneous Chinese liquor fermentation. Appl Environ Microbiol 83:e01475-17. doi: 10.1128/AEM.01475-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardinale M, Grube M, Erlacher A, Quehenberger J, Berg G. 2015. Bacterial networks and cooccurrence relationships in the lettuce root microbiota. Environ Microbiol 17:239–252. doi: 10.1111/1462-2920.12686. [DOI] [PubMed] [Google Scholar]
- 18.Parente E, Cocolin L, De Filippis F, Zotta T, Ferrocino I, O’Sullivan O, Neviani E, De Angelis M, Cotter PD, Ercolini D. 2016. FoodMicrobionet: a database for the visualisation and exploration of food bacterial communities based on network analysis. Int J Food Microbiol 219:28–37. doi: 10.1016/j.ijfoodmicro.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Jin GY, Zhu Y, Xu Y. 2017. Mystery behind Chinese liquor fermentation. Trends Food Sci Technol 63:18–28. doi: 10.1016/j.tifs.2017.02.016. [DOI] [Google Scholar]
- 20.Zhang CL, Ao ZH, Chui WQ, Shen CH, Tao WY, Zhang SY. 2012. Characterization of the aroma-active compounds in daqu: a tradition Chinese liquor starter. Eur Food Res Technol 234:69–76. doi: 10.1007/s00217-011-1616-4. [DOI] [Google Scholar]
- 21.Lemos LN, Fulthorpe RR, Triplett EW, Roesch LF. 2011. Rethinking microbial diversity analysis in the high throughput sequencing era. J Microbiol Methods 86:42–51. doi: 10.1016/j.mimet.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Du H, Xu Y. 2017. Source tracking of prokaryotic communities in fermented grain of Chinese strong-flavor liquor. Int J Food Microbiol 244:27–35. doi: 10.1016/j.ijfoodmicro.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 23.Rui JP, Li JB, Zhang SH, Yan XF, Wang YP, Li XZ. 2015. The core populations and cooccurrence patterns of prokaryotic communities in household biogas digesters. Biotechnol Biofuels 8:158. doi: 10.1186/s13068-015-0339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao WJ, Fan WL, Xu Y. 2014. Characterization of the key odorants in light aroma type Chinese liquor by gas chromatography—olfactometry, quantitative measurements, aroma recombination, and omission studies. J Agric Food Chem 62:5796–5804. doi: 10.1021/jf501214c. [DOI] [PubMed] [Google Scholar]
- 25.Cao Y, Hu Y. 2010. Variation of aromatic components in solid phase fermented grains during fermentation of fen liquor. Food Sci 31:367–371. [Google Scholar]
- 26.Barberán A, Bates ST, Casamayor EO, Fierer N. 2012. Using network analysis to explore cooccurrence patterns in soil microbial communities. ISME J 6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Filippis F, Genovese A, Ferranti P, Gilbert JA, Ercolini D. 2016. Metatranscriptomics reveals temperature-driven functional changes in microbiome impacting cheese maturation rate. Sci Rep 6:21871. doi: 10.1038/srep21871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinhauser D, Krall L, Müssig C, Büssis D, Usadel B. 2007. Correlation networks In Junker BH, Schreiber F (ed), Analysis of biological networks. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- 29.Yang J, Leskovec J. 2014. Overlapping communities explain core-periphery organization of networks. Proc IEEE 102:1892–1902. doi: 10.1109/JPROC.2014.2364018. [DOI] [Google Scholar]
- 30.Ercolini D, Pontonio E, Filippis FD, Minervini F, Storia AL, Gobbetti M, Cagno RD. 2013. Microbial ecology dynamics during rye and wheat sourdough preparation. Appl Environ Microbiol 79:7827–7836. doi: 10.1128/AEM.02955-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, Gao Y, Wang S, Xu D, Yu H, Wu L, Lin Q, Hu Y, Li X, He Z, Deng Y, Zhou J. 2014. The microbial gene diversity along an elevation gradient of the Tibetan grassland. ISME J 8:430. doi: 10.1038/ismej.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Bruyn F, Zhang SJ, Pothakos V, Torres J, Lambot C, Moroni AV, Callanan M, Sybesma W, Weckx S, De Vuyst L. 2017. Exploring the impacts of postharvest processing on the microbiota and metabolite profiles during green coffee bean production. Appl Environ Microbiol 83:e02398-16. doi: 10.1128/AEM.02398-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang J, Tang X, Tang M, Zhang X, Xu X, Yi Y. 2017. Analysis of the bacterial communities in two liquors of soy sauce aroma as revealed by high-throughput sequencing of the 16S rRNA V4 hypervariable region. Biomed Res Int 2017:6271358. doi: 10.1155/2017/6271358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X, Teng K, Zhang J, Wang F, Zhang T, Ai G, Han P, Bai F, Zhong J. 2017. Transcriptome responses of Lactobacillus acetotolerans F28 to a short and long term ethanol stress. Sci Rep 7:2650. doi: 10.1038/s41598-017-02975-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toh H, Morita H, Tsuji H, Iwashita K, Goto N, Nakayama J, Sekine M, Kato Y, Suzuki K, Fujita N. 2015. Complete genome sequence of Lactobacillus acetotolerans RIB 9124 (NBRC 13120) isolated from putrefied (hiochi) Japanese sake. J Biotechnol 214:214–215. doi: 10.1016/j.jbiotec.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Wu Q, Zhu W, Wang W, Xu Y. 2015. Effect of yeast species on the terpenoids profile of Chinese light-style liquor. Food Chem 168:390–395. doi: 10.1016/j.foodchem.2014.07.069. [DOI] [PubMed] [Google Scholar]
- 37.Tian X, Cui H, Wang X. 2016. Research progress on the application of the fermentation products of Geotrichum candidum. Anhui Agric Sci Bull 22:31–32. [Google Scholar]
- 38.Noronha-da-Costa P, Rodrigues C, Spencer-Martins I, Loureiro V. 1996. Fatty acid patterns of film-forming yeasts and new evidence for the heterogeneity of Pichia membranaefaciens. Lett Appl Microbiol 23:79–84. doi: 10.1111/j.1472-765X.1996.tb00035.x. [DOI] [PubMed] [Google Scholar]
- 39.Gong GL, Ma LY, Chen XF. 2014. Isolation and improvement of Saccharomyces cerevisiae for producing the distilled liquor. J Chem Pharm Res 6:283–288. [Google Scholar]
- 40.Kong Y, Wu Q, Zhang Y, Xu Y. 2014. In situ analysis of metabolic characteristics reveals the key yeast in the spontaneous and solid-state fermentation process of Chinese light-style liquor. Appl Environ Microbiol 80:3667–3676. doi: 10.1128/AEM.04219-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu XL, Wang HY, Wu Q, Xu Y. 2014. Development, validation and application of specific primers for analyzing the clostridial diversity in dark fermentation pit mud by PCR-DGGE. Bioresour Technol 163:40–47. doi: 10.1016/j.biortech.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 42.Wang P, Wu Q, Jiang X, Wang Z, Tang J, Xu Y. 2017. Bacillus licheniformis affects the microbial community and metabolic profile in the spontaneous fermentation of daqu starter for Chinese liquor making. Int J Food Microbiol 250:59–67. doi: 10.1016/j.ijfoodmicro.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 43.Niu Y, Yao Z, Xiao Q, Xiao Z, Ma N, Zhu J. 2017. Characterization of the key aroma compounds in different light aroma type Chinese liquors by GC-olfactometry, GC-FPD, quantitative measurements, and aroma recombination. Food Chem 233:204–215. doi: 10.1016/j.foodchem.2017.04.103. [DOI] [PubMed] [Google Scholar]
- 44.Monnet C, Dugat-Bony E, Swennen D, Beckerich J-M, Irlinger F, Fraud S, Bonnarme P. 2016. Investigation of the activity of the microorganisms in a Reblochon-style cheese by metatranscriptomic analysis. Front Microbiol 7:536. doi: 10.3389/fmicb.2016.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li S, Li P, Liu X, Luo LX, Lin WF. 2016. Bacterial dynamics and metabolite changes in solid-state acetic acid fermentation of Shanxi aged vinegar. Appl Microbiol Biotechnol 100:4395–4411. doi: 10.1007/s00253-016-7284-3. [DOI] [PubMed] [Google Scholar]
- 46.Kable ME, Srisengfa Y, Laird M, Zaragoza J, McLeod J, Heidenreich J, Marco ML. 2016. The core and seasonal microbiota of raw bovine milk in tanker trucks and the impact of transfer to a milk processing facility. mBio 7:e00836-16. doi: 10.1128/mBio.00836-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Annan NT, Poll L, Sefa-Dedeh S, Plahar WA, Jakobsen M. 2003. Volatile compounds produced by Lactobacillus fermentum, Saccharomyces cerevisiae and Candida krusei in single starter culture fermentations of Ghanaian maize dough. J Appl Microbiol 94:462–474. doi: 10.1046/j.1365-2672.2003.01852.x. [DOI] [PubMed] [Google Scholar]
- 48.Laakso K, Koskenniemi K, Koponen J, Kankainen M, Surakka A, Salusjärvi T, Auvinen P, Savijoki K, Nyman TA, Kalkkinen N, Tynkkynen S, Varmanen P. 2011. Growth phase-associated changes in the proteome and transcriptome of Lactobacillus rhamnosus GG in industrial-type whey medium. Microb Biotechnol 4:746–766. doi: 10.1111/j.1751-7915.2011.00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang PX, Mao J, Meng XY, Li XZ, Liu YY, Feng H. 2014. Changes in flavour characteristics and bacterial diversity during the traditional fermentation of Chinese rice wines from Shaoxing region. Food Control 44:58–63. doi: 10.1016/j.foodcont.2014.03.018. [DOI] [Google Scholar]
- 50.Li P, Li S, Cheng L, Luo L. 2014. Analyzing the relation between the microbial diversity of Daqu and the turbidity spoilage of traditional Chinese vinegar. Appl Microbiol Biotechnol 98:6073–6084. doi: 10.1007/s00253-014-5697-4. [DOI] [PubMed] [Google Scholar]
- 51.Almeida M, Hébert A, Abraham A-L, Rasmussen S, Monnet C, Pons N, Delbès C, Loux V, Batto J-M, Leonard P, Kennedy S, Ehrlich SD, Pop M, Montel M-C, Irlinger F, Renault P. 2014. Construction of a dairy microbial genome catalog opens new perspectives for the metagenomic analysis of dairy fermented products. BMC Genomics 15:1101–1116. doi: 10.1186/1471-2164-15-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balat M. 2011. Production of bioethanol from lignocellulosic materials via the biochemical pathway: a review. Energy Convers Manag 52:858–875. doi: 10.1016/j.enconman.2010.08.013. [DOI] [Google Scholar]
- 53.Mattanovich D, Sauer M, Gasser B. 2016. Industrial microorganisms: Pichia pastoris, p 687–714. In Wittmann C, Liao JC (ed), Industrial biotechnology: microorganisms, vol 1 Wiley-Blackwell, Hoboken, NJ. [Google Scholar]
- 54.Reference deleted.
- 55.Rojas V, Gil JV, Piñaga F, Manzanares P. 2001. Studies on acetate ester production by non-Saccharomyces wine yeasts. Int J Food Microbiol 70:283–289. doi: 10.1016/S0168-1605(01)00552-9. [DOI] [PubMed] [Google Scholar]
- 56.Veeraragavan K, Colpitts T, Gibbs B. 1990. Purification and characterization of 2 distinct lipases from Geotrichum candidum. Biochim Biophys Acta 1044:26–33. doi: 10.1016/0005-2760(90)90214-I. [DOI] [PubMed] [Google Scholar]
- 57.Burkert JF, Maugeri F, Rodrigues MI. 2004. Optimization of extracellular lipase production by Geotrichum sp. using factorial design. Bioresour Technol 91:77–84. doi: 10.1016/S0960-8524(03)00152-4. [DOI] [PubMed] [Google Scholar]
- 58.Sukpipat W, Komeda H, Prasertsan P, Asano Y. 2017. Purification and characterization of xylitol dehydrogenase with l -arabitol dehydrogenase activity from the newly isolated pentose-fermenting yeast Meyerozyma caribbica 5XY2. J Biosci Bioeng 123:20–27. doi: 10.1016/j.jbiosc.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 59.Bourrie BC, Willing BP, Cotter PD. 2016. The microbiota and health promoting characteristics of the fermented beverage kefir. Front Microbiol 7:674. doi: 10.3389/fmicb.2016.00647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gethins L, Guneser O, Demirkol A, Rea MC, Stanton C, Ross RP, Yuceer Y, Morrissey JP. 2015. Influence of carbon and nitrogen source on production of volatile fragrance and flavour metabolites by the yeast Kluyveromyces marxianus. Yeast 32:67–76. doi: 10.1002/yea.3047. [DOI] [PubMed] [Google Scholar]
- 61.Ren K, Lamsal BP. 2017. Synthesis of some glucose-fatty acid esters by lipase from Candida antarctica and their emulsion functions. Food Chem 214:556–563. doi: 10.1016/j.foodchem.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 62.Trbojević Ivić J, Milosavić N, Dimitrijević A, Jankulović MG, Bezbradica D, Kolarski D, Veličković D. 2017. Synthesis of medium-chain length capsinoids from coconut oil catalyzed by Candida rugosa lipases. Food Chem 218:505–508. doi: 10.1016/j.foodchem.2016.09.049. [DOI] [PubMed] [Google Scholar]
- 63.He Y, Li J, Kodali S, Chen B, Guo Z. 2017. Rationale behind the near-ideal catalysis of Candida antarctica lipase A (CAL-A) for highly concentrating ω-3 polyunsaturated fatty acids into monoacylglycerols. Food Chem 219:230–239. doi: 10.1016/j.foodchem.2016.09.149. [DOI] [PubMed] [Google Scholar]
- 64.Englezos V, Torchio F, Cravero F, Marengo F, Giacosa S, Gerbi V, Rantsiou K, Rolle L, Cocolin L. 2016. Aroma profile and composition of Barbera wines obtained by mixed fermentations of Starmerella bacillaris (synonym Candida zemplinina) and Saccharomyces cerevisiae. LWT 73:567–575. doi: 10.1016/j.lwt.2016.06.063. [DOI] [Google Scholar]
- 65.Zhang XL, Tian XQ, Ma LY, Feng B, Liu QH, Yuan LD, Fan CQ, Huang HL, Yang Q. 2015. Biodiversity of the symbiotic bacteria associated with toxic marine dinoflagellate Alexandrium tamarense. J Biosci Med 3:57105. doi: 10.4236/jbm.2015.36004. [DOI] [Google Scholar]
- 66.Toju H, Tanabe AS, Yamamoto S, Sato H. 2012. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS One 7:e40863. doi: 10.1371/journal.pone.0040863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ren G, Ren W, Teng Y, Li Z. 2015. Evident bacterial community changes but only slight degradation when polluted with pyrene in a red soil. Front Microbiol 6:22. doi: 10.3389/fmicb.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hierro N, Esteve-Zarzoso B, González Á, Mas A, Guillamón JM. 2006. Real-time quantitative PCR (qPCR) and reverse transcription-qPCR for detection and enumeration of total yeasts in wine. Appl Environ Microbiol 72:7148–7155. doi: 10.1128/AEM.00388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mayrhofer S, Filipp R, Lehner D, Reiterich C, Kneifel W, Domig KJ. 2016. Suitability of different PCR-DGGE primer sets for the monitoring of lactic acid bacteria in wine. S Afr J Enol Vitic 35:185–195. [Google Scholar]
- 70.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 73.Tao Y, Li J, Rui J, Xu Z, Zhou Y, Hu X, Wang X, Liu M, Li D, Li X. 2014. Prokaryotic communities in pit mud from different-aged cellars used for the production of Chinese strong-flavored liquor. Appl Environ Microbiol 80:2254–2260. doi: 10.1128/AEM.04070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 75.Li P, Lin W, Liu X, Wang X, Gan X, Luo L, Lin WT. 2017. Effect of bioaugmented inoculation on microbiota dynamics during solid-state fermentation of daqu starter using autochthonous of Bacillus, Pediococcus, Wickerhamomyces and Saccharomycopsis. Food Microbiol 61:83–92. doi: 10.1016/j.fm.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 76.Wu Q, Chen L, Xu Y. 2013. Yeast community associated with the solid state fermentation of traditional Chinese Maotai-flavor liquor. Int J Food Microbiol 166:323–330. doi: 10.1016/j.ijfoodmicro.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 77.Wu Q, Kong Y, Xu Y. 2016. Flavor profile of Chinese liquor is altered by interactions of intrinsic and extrinsic microbes. Appl Environ Microbiol 82:422–430. doi: 10.1128/AEM.02518-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ, Suggests M. 2008. The vegan package. http://vegan.r-forge.r-project.org/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The fungal and bacterial raw sequence data were deposited in the DNA Data Bank of Japan (DDBJ) database under the accession numbers DRA005471 and DRA005474, respectively.