Figure 2.
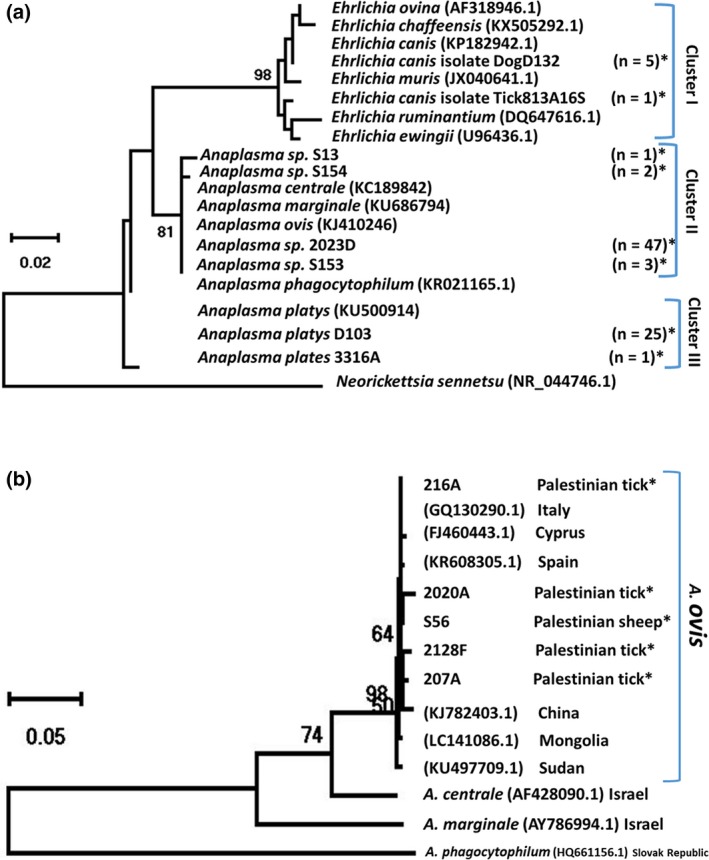
Phylogenetic analysis of Anaplasma and Ehrlichia based on partial sequences of (a) 16S rRNA and (b) msp4 genes. Phylogenetic analysis were constructed using the maximum likelihood method used on MEGA X program, with the complete deletion option, based on Jukes‐Cantor model for nucleotide sequences. Initial trees for the heuristic search were automatically obtained by applying the Nearest‐Neighbor‐Interchange (NNI) algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach. Statistical support for internal branches of the trees was evaluated using bootstrap of 1000 replications. (a) Based on 16srRNA sequences (345 bp): cluster I represents Ehrlichia strains. Cluster II represents Anaplasma spp. and Cluster III represents Anaplasma platys. The DNA sequence of Neorickettsia sennetsu (NR_044746.1) was used as an out‐group to produce rooted tree. (b): 851 bp msp4 DNA Anaplasma sequences detected in this study compared to Anaplasma reference sequences deposited in the NCBI GenBank. The Species, GenBank accession numbers and country of origin from which the sequences were derived are included for each sequence. Sequences derived from this study are marked by star (*). The number of identical sequences is in brackets. Selected reference Anaplasma spp. sequences from GenBank are also shown.
