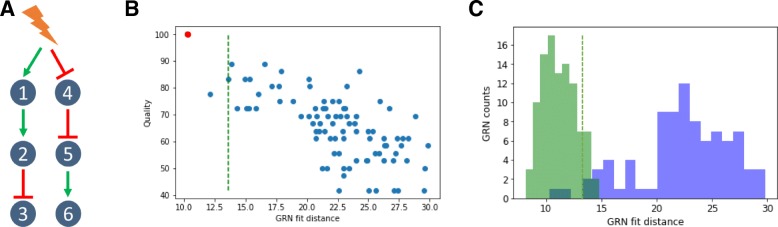
Fig. 3.
In silico cascade GRN inference a The cascade GRN. Genes parameters were taken from in vitro estimations to mimic realistic behavior. Experimental data were generated to obtain time courses of transciptomic data, at single-cell and population scale, and also proteomic data at population scale. b WASABI was run to infer in silico cascade GRN and generated 88 candidates. A dot represents a network candidate with its associated fit distance and inference quality (percentage of true interactions). True GRN is inferred (red dot, 100% quality). Acceptable maximum fit distance (green dashed line) corresponds to variability of true GRN fit distance. Its computation is detailed in figure C. Three GRN candidates (including the true one) have a fit distance below threshold. c Variability of true GRN fit distance (green dashed line in figures B and C) is estimated as the threshold where 95% of true GRN fit distance is below. Fit distance distribution is represented for true GRN (green) and candidates (blue) for cascade in silico GRN benchmark. True GRNs are calibrated by WASABI directed inference while candidates are inferred from non-directed inference. Fit distance represents similitude between candidates generated data and reference experimental data