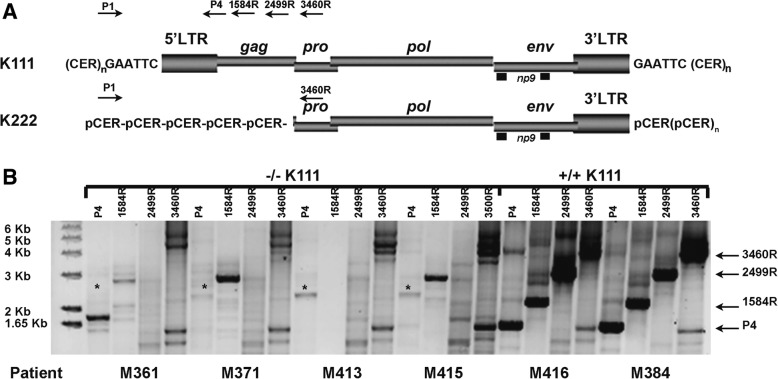
Fig. 2.
5′ Mapping of pericentromeric HERV-K proviruses in CTCL patients. a Schematic representation of the primer sets used to amplify centromeric HERV-K (HML-2) proviruses by PCR. The genomic structure of centromeric HERV-K (HML-2) proviruses K111 and K222 is shown; the viral genes gag, pro, pol, env and np9, surrounded by LTRs, are integrated into centromeric repeats (CER:D22Z3). At the time of integration, target site duplications are produced at each site of the provirus, which in the case of K111 is GAATTC. The forward primer P1 was used in combination with four reverse primers: P4, 1584, 2499 and 3460 (the numbers correspond to the nucleotide position in reference to K111) that span the HERV-K (HML-2) gag and pro genes. The product of these primer sets is indicated by the arrows on the right i.e. P4 etc. b K222 provirus is detected by PCR in the DNA of the CTCL −/−K111 patients only by the primer set P1–3460 (low molecular weight band shown by the arrow), which was confirmed by sequencing [26]. These DNAs do not produce the expected bands obtained with the 3 reverse primers P4, 1584 and 2499 as shown in the samples from patients with a +/+K111 genotype. The asterisk indicates a band, shown by sequencing, to be a non-specific PCR amplification product of HERV-K (HML-2) 3p25.3