Abstract
Mutations in the gene encoding epidermal growth factor receptor (EGFR) family member HER2 (ERBB2) are common in and drive the growth of “HER2-negative” (not ERBB2-amplified) solid tumors but are rare in primary “HER2-positive” (ERBB2-amplified) breast cancer. We analyzed DNA sequencing data from HER2-positive patients and used cell lines and a patient-derived xenograft model to test the effects of HER2 mutations on the efficacy of anti-HER2 agents such as trastuzumab, lapatinib, or the irreversible pan-EGFR inhibitor neratinib. HER2 mutations were present in ~ 7% of HER2-positive tumors, all of which were metastatic but not all were previously treated. Compared to HER2-amplification alone, in both patients and cultured cell lines, co-occurrence of HER2 mutation and amplification was associated with poor response to trastuzumab and lapatinib, standard-of-care anti-HER2 agents. In mice, xenografts established from a patient whose HER2-positive tumor acquired a D769Y mutation in HER2 after progression on trastuzumab-based therapy were resistant to trastuzumab or lapatinib but were sensitive to neratinib. Clinical data revealed that six heavily pretreated patients with tumors bearing coincident HER2 amplification and mutation subsequently exhibited significant response to neratinib monotherapy. Thus, the findings indicate that coincident HER2 mutation reduces the efficacy of therapies commonly used to treat HER2-positive breast cancer, particularly in metastatic and previously HER2-inhibitor–treated patients as well as potentially in patients scheduled for first-line treatment. We propose, therefore, that clinical studies testing the efficacy of neratinib are warranted selectively in breast cancer patients whose tumors carry both amplification and mutation of HER2/ERBB2.
Introduction
Overexpression of human epidermal growth factor receptor 2 (HER2), usually because of amplification of its gene ERBB2, occurs in ~20% of breast cancer patients. Before the development of specific HER2-targeted agents such as trastuzumab and lapatinib, and later pertuzumab and T-DM1 [reviewed in (1)], patients with “HER2-positive” (ERBB2-amplified; hereafter HER2-amplified) breast cancer had an extremely poor prognosis (2, 3). Thanks to these therapeutic options, HER2-positive breast cancer is now considered a highly curable disease when diagnosed before metastatic spread. On the contrary, patients with advanced disease are frequently refractory to the therapy or, in patients initially sensitive to these agents, acquired resistance inevitably emerges over time.
Besides gene amplification, the kinase activity of HER2 can increase as a result of activating mutations in the extracellular, juxtamembrane, or intracellular regions (4, 5). The perturbation in HER2’s catalytic activity triggered by these mutations and their intrinsic sensitivity to small-molecule HER family kinase inhibitors has been studied in several preclinical models of “HER2-negative” (HER2 expressed but not amplified) cancers by overexpressing the mutant isoforms of the receptor (5–8). Increasing efforts in deep sequencing of tumors revealed several of these mutations in various HER2-negative cancers (9, 10). These premises were the basis for a basket trial in which the efficacy of the pan-HER kinase inhibitor neratinib was tested in solid tumors bearing HER2 mutations (11). In that study, we showed that HER2 kinase inhibition has antitumor activity in this setting, with breast cancer patients being the most sensitive among the enrollees.
It is generally accepted that the presence of HER2 mutations in breast cancer is mutually exclusive with amplification of the receptor. This notion is largely based on genome sequencing efforts conducted mainly in primary tumors. In the TCGA database, for example, only 3 cases out of 119 HER2-amplifed breast cancer patients (2.5%) show co-occurrence of HER2 mutations (12). Similarly, in the METABRIC cohort (13), only nine out 342 HER2-amplified patients (2.6%) also bear HER2 mutations. In this study, we investigated the frequency of HER2 mutation and amplification co-occurrence in breast cancer and assess the antitumor activity of the HER tyrosine kinase inhibitor neratinib in preclinical models and patients with coincident HER2-mutant/HER2-amplified breast cancer.
Results
The presence of ERBB2 mutations in HER2-positive breast cancer correlates with poor response to standard-of-care anti-HER2 therapy
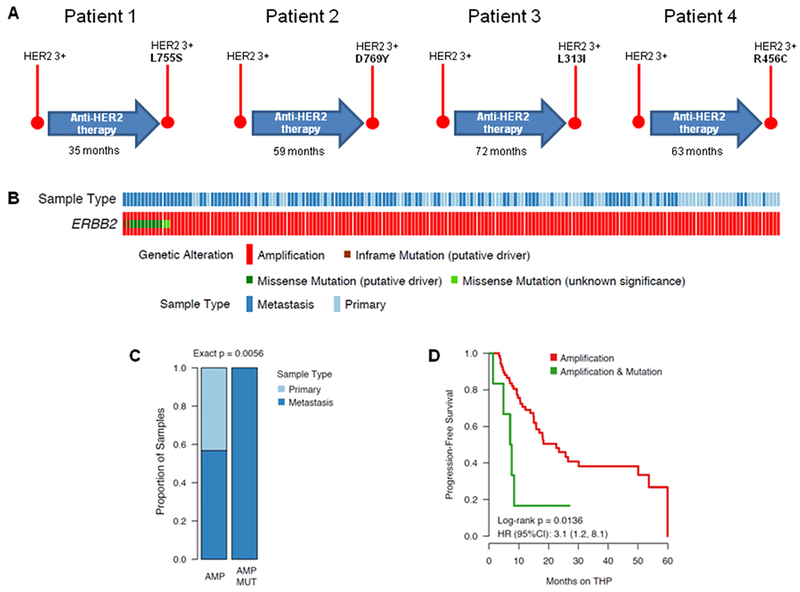
As part of our efforts to sequence HER2-positive tumors relapsing to standard-of-care HER2 blockade, we observed that, out of a cohort of 77 patients with HER2-positive breast cancer (33 patients treated at START Madrid, 23 patients treated at Peter MacCallum Cancer Centre and 20 patients treated at Memorial Sloan Kettering Cancer Center), the tumors from four of these patients treated with one of several lines of standard-of-care anti-HER2 agents (trastuzumab, lapatinib, and/or a trastuzumab-emtansine antibody-drug conjugate known as T-DM1) acquired a HER2 mutation at time of disease progression (Fig. 1A and text S1). Specifically, one patient treated for 35 months with trastuzumab or lapatinib-based therapy developed a HER2 L755S mutation at progression, one patient acquired a HER2 D769Y mutation after 59 months of therapy with trastuzumab, lapatinib, and T-DM1, and the remaining two patients acquired extracellular domain HER2 L313I or R456C mutations after 72 and 63 months of anti-HER2 therapy, respectively. The details of the clinical histories of these patients are in the supplementary materials (text S1), but a common denominator of these four cases is the metastatic nature of their disease. The somatic HER2 L755S mutation is the most frequent in breast cancer, and was previously described to limit the activity of lapatinib in cultured cells (5, 8). Likewise, the HER2 D769Y mutation is a well-characterized activating mutation as studied in nonmalignant mammary epithelial MCF10A cells (5). Mutations of the Arg456 amino acid and the Leu313 residue (close to the HER2 Ser310 hotspot) are less common. The R456C mutation was previously detected in a prostate adenocarcinoma sequenced at MSKCC, while mutations in the Leu313 residue were detected in a bladder carcinoma and endocervical adenocarcinoma in our MSKCC clinical sequencing cohort. Notably, in the bladder carcinoma, the L313V mutation was accompanied by the amplification of the gene.
Fig. 1: Co-existence of ERBB2 amplification and mutations in metastatic breast cancer.
(A) Schematic representation of the detection of HER2 mutations in HER2-positive breast cancer patients treated with anti-HER2 therapy for the indicated time. “HER2 3+” refers to tumors that were highly positive for HER2 staining by IHC. (B) OncoPrint software-generated genomic alterations heatmap of the ERBB2-amplified patients available in the MSK database. (C) Graph indicating the proportion of primary or metastatic patients with tumors bearing concomitant mutation and amplification of ERBB2. (D) Kaplen-Meyer curves showing the progression-free survival of the MSK cohort of HER2-positive patients treated with the standard-of-care trastuzumab/pertuzumab/paclitaxel combination (THP). Displayed inset is the hazard ratio (HR) with 95% confidence interval (95%CI) of patients with ERBB2-mutant/amplified (coincident) tumors versus ERBB2-amplified–only tumors.
These isolated cases prompted us to investigate the prevalence of HER2 mutations among a larger cohort of HER2-positive breast cancer patients sequenced by the MSK-IMPACT (Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets) DNA-sequencing platform. Out of 184 patients with detected HER2 amplification by MSK-IMPACT, 13 cases (7.1%) had concomitant mutations of the receptor (Fig. 1B). Notably, all of these 13 tumors were metastatic, whereas only 56.7% (97/171) of the tumors with HER2 amplification-alone were metastatic (Fisher exact P = 0.0056; Fig. 1C). We then evaluated whether the enrichment of HER2-amplified/mutant tumors that we observed in the metastatic setting could be a consequence of therapy selection rather than the metastatic status of the disease. To do so, we analyzed our cohort to evaluate the rate of ERBB2 mutation-amplification and amplification alone comparing the treatment-naïve vs post-treatment specimens. The results showed that the mutation and amplification is indeed more prevalent in post-treatment samples than in treatment-naïve tumors (MUT-AMP: 11/13 [85%] vs AMP: 74/171[43%]. However, this analysis was insufficiently powered to identify robust differences between the two groups possibly due to the small numbers of patients with concomitant mutated and amplified tumors (Exact p = 0.076; fig. S1). Coincident HER2 mutation and amplification was also associated with poor response to a trastuzumab/pertuzumab/paclitaxel combination therapy compared to HER2 amplification alone [on the basis of progression-free survival, which was 7.3 months in the coincident group (95% confidence interval: 4.8, not reached) and 22.6 months in the amplification-alone group (95% confidence interval: 15.8, 53.7), log-rank P = 0.0136; Fig. 1D]. Notably, further adjustment for sample type (metastatic vs primary) or line of therapy in the metastatic setting did not change these results.
Exogenous expression of a HER2 mutation limits the sensitivity to trastuzumab and lapatinib in HER2 amplified breast cancer cells
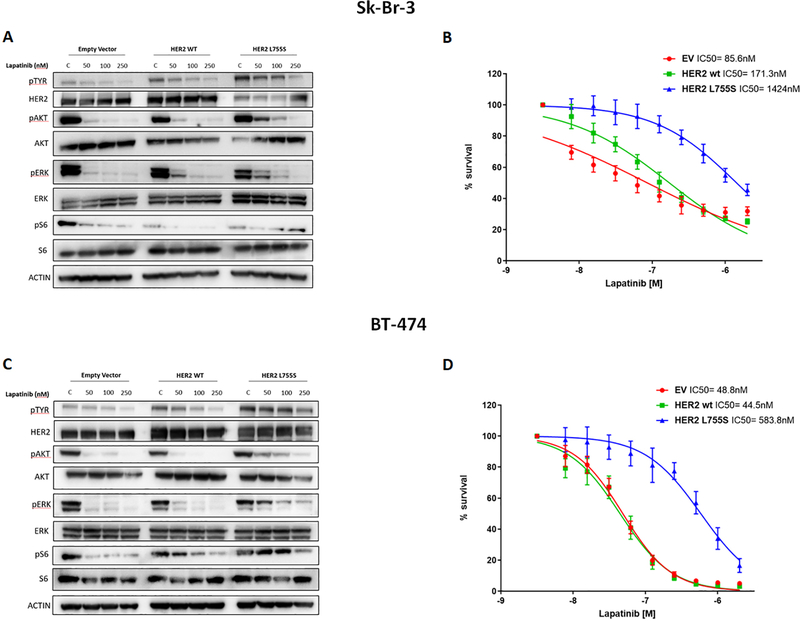
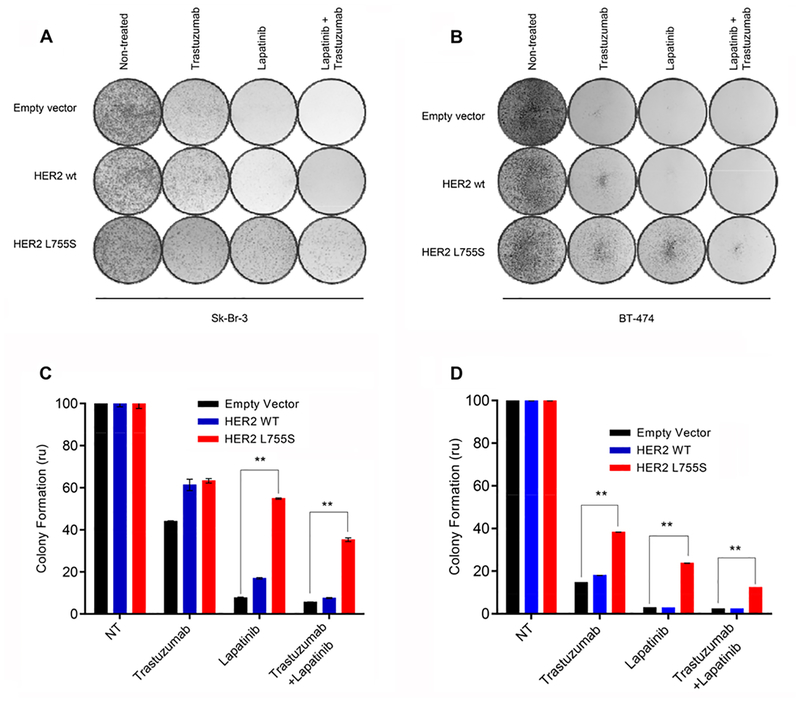
Our results, together with both preclinical (14) and clinical (15) observations recently reported, suggest that the acquisition of HER2 mutation in the metastatic setting may affect the sensitivity to anti-HER2 therapy in HER2-positive breast cancer patients. To confirm that the presence of HER2 mutations is sufficient to limit the sensitivity to trastuzumab or lapatinib in a HER2-amplified background, we stably transduced Sk-Br-3 and BT474 cells with lentiviral vectors encoding for wild-type or mutant (L755S) HER2 (or an empty vector control). We chose the L755S mutation because is the most frequently found in breast cancer and was detected in one of the 4 patients treated with anti-HER2 therapy described above. Treatment with lapatinib markedly suppressed the phosphorylation of HER2 downstream signaling proteins, namely the kinases AKT and extracellular signal-regulated kinase (ERK) and ribosomal protein S6, in both wild-type– and empty-vector–transduced cells. The same concentrations of lapatinib were found to be less effective in inhibiting AKT, ERK and S6 signaling in the HER2 L755S-transduced cells (Fig. 2, A and C). Consequently, lapatinib had significantly less anti-proliferative activity in Sk-Br-3 and BT474 cells expressing the L755S HER2 mutation compared to cells expressing wild-type HER2 (IC50= 1424 nM vs 171.3 nM and 583.8 nM vs 44.5 nM for the Sk-Br-3 and the BT474 clones, respectively; P<0.05; Fig. 2, B and D).Consistently, expression of the L755S HER2 variant was sufficient to limit the sensitivity of these cell lines also to trastuzumab or the combination of trastuzumab and lapatinib, as compared with wild-type HER2–transfected or empty-vector–transfected cells in long-term clonogenic assays (Fig. 3, A to D).
Fig. 2. HER2 L755S mutation confers resistance to lapatinib.
(A and C) Western blot analyses of total HER2, AKT, ERK and S6 as well as phospho-Tyrosine, phospho AKT (Ser473), phospho ERK (Thr202/Tyr204 and phospho S6 (Ser235/236) in whole-cell extracts from Sk-Br-3 (A) and BT-474 (C) cells expressing empty-vector, wild-type (WT) HER2, or HER2 L755S and cultured in various concentrations of lapatinib for 4 hours. (p, phosphorylated). (B and D). Cell viability by CTG assay of the cells described in (A) and (C), respectively after incubation with increasing concentrations of lapatinib (ranging from 4 to 2000nM). Data are means ± SD from three independent experiments. P value obtained by two-tailed Student’s t-test.
Fig. 3. HER2 L755S mutation induces resistance to HER2-targeted therapy.
Clonogenic growth assays performed by crystal violet staining on Sk-Br-3 (A) and BT-474 (B) cells expressing empty-vector, wild-type or mutant (L755S) HER2 constructs and cultured with trastuzumab (20 μg/ml), lapatinib (500 nM) or the combination, as indicated, for ten days. Results from treated cultures were quantified relative to non-treated cells. Data are means ± SEM from three independent experiments. *P<0.05 by two-tailed Student’s t-test.
Efficacy of neratinib in breast cancer cells with coincident amplification and mutation of HER2
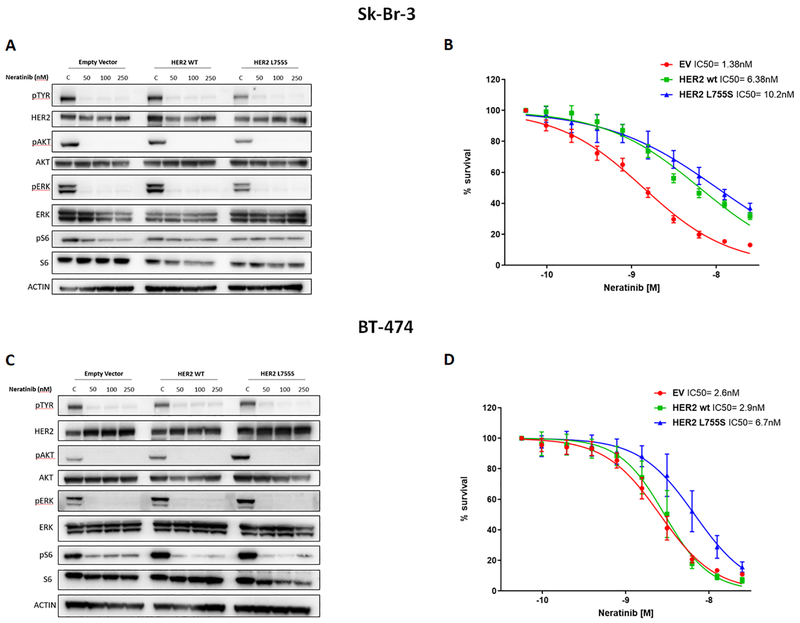
Recent reports have shown that neratinib, an irreversible pan-HER tyrosine kinase inhibitor, can suppress the proliferation and tumorigenesis of HER2-negative cells engineered to overexpress mutant isoforms of HER2 (5). Thus, we reasoned that neratinib would inhibit the growth of tumors bearing both amplification and mutations of ERBB2. To test this hypothesis, we evaluated the effects of neratinib treatment on the phosphorylation status of receptor tyrosine kinases (RTKs) and HER2 downstream signaling proteins in the empty vector, HER2 WT and HER2 L755S-transduced Sk-Br-3 and BT474-derived cells. We found that neratinib was able to equally inhibit RTKs phosphorylation and HER2 downstream signaling proteins in all the transduced cell lines (Fig. 4 A and C). Consistently, proliferation assays showed that neratinib was highly efficient in inhibiting the growth of the HER2 L755S-trasduced cells with IC50 in the low nanomolar range (IC50= 10.2nM and 6.7nM for the HER2 L755S-transduced Sk-Br-3 and BT474 cells, respectively; Fig. 4 B and D). Together these data suggest that neratinib is more potent than lapatinib against breast cancers harboring both HER2 amplification and mutation.
Fig. 4. Activity of neratinib against breast cancer cells carrying both the amplification and mutation of HER2.
(A and C) Western blot analyses of total HER2, AKT, ERK and S6 as well as phospho-Tyrosine, phospho AKT (Ser473), phospho ERK (Thr202/Tyr204) and phospho S6 (Ser235/236) in whole-cell extracts from Sk-Br-3 (A) and BT-474 (C) cells expressing empty-vector, wild-type (WT) HER2, or HER2 L755S and cultured in various concentrations of neratinib for 4 hours. (p, phosphorylated). (B and D) Cell viability by CTG assay of the cells described in (A) and (C), respectively after incubation with increasing concentrations of lapatinib (ranging from 0.05 to 25nM). Data are means ± SD from three independent experiments. P value obtained by two-tailed Student’s t-test.
In vivo efficacy of neratinib
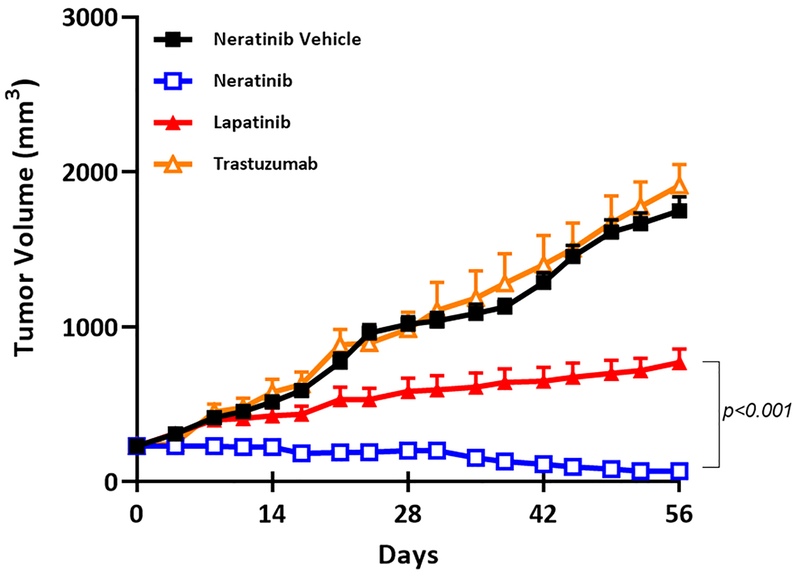
To test the efficacy of neratinib in vivo against breast tumors with coincident amplification and mutation of HER2, we established sub-cutaneous patient-derived xenografts (PDXs) from a HER2-positive breast cancer patient whose metastatic lung lesions carried both a D769Y mutation and amplification of HER2. Mice were treated with vehicle, trastuzumab, lapatinib, or neratinib for just under 5 weeks. As expected, xenografts were refractory to both trastuzumab and lapatinib (Fig. 5), suggesting that the presence of this HER2 mutation may lead to cross-resistance to multiple anti-HER2 agents. Conversely, neratinib significantly inhibited tumor growth and, in enough cases to have a cohort-level effect, neratinib induced durable tumor shrinkage (Fig. 5).
Fig. 5. In vivo efficacy of neratinib against HER2-amplified and mutant PDXs.
Antitumor effects of neratinib in xenografts bearing in ERBB2. Growth of patient-derived xenografts containing coincident ERBB2 amplification and HER2 D769Y mutation, in mice treated with vehicle (control), trastuzumab (10 mg/kg; ip; 2xwkly), lapatinib (100mg/kg daily) or neratinib (40mg/kg daily). Data are k means ± SEM from 8 mice each condition. P<0.001 by two-tailed Student’s t-test
Neratinib has clinical activity in breast cancer patients with tumors bearing ERBB2 amplification and mutations
Our findings indicate that the acquisition of HER2 mutations confer resistance to current standard anti-HER2 therapies but not to neratinib. Moreover, we have recently shown that neratinib has clinical activity in breast cancer patients with HER2-negative tumors bearing HER2 mutations (11). Thus, we posited that neratinib would be effective in tumors with co-existing HER2 amplification and mutation. We tested this hypothesis in six breast cancer patients with these characteristics (Table 1). Patients A-C (clinical details in text S1) were enrolled as part of the breast cancer cohort of the SUMMIT “basket” trial that was recently published (9); these patients received 240 mg/day neratinib monotherapy. Patients 2–4 (described in Fig. 1A and text S1) received neratinib as compassionate use. Notably, four patients achieved stabilization of the disease (from 4 up to 7 months), and two patients experienced significant reduction of tumor volume, in one case lasting up to 10 months. Together, these results show that neratinib has clinical activity in breast tumors carrying both amplification and mutations of HER2/ERBB2.
Table 1. Clinical benefit of neratinib treatment in HER2-amplified/mutant breast cancer patients.
SD, stable disease (tumor volume changing within the +20% or −30% range compared to baseline pre-treatment); PR, partial response (tumor volume decreasing more than 30% compared to baseline pre-treatment).
| Patient | HER2 mutation |
ERBB2 gene copies |
Best response to neratinib |
Time on treatment (days) |
|---|---|---|---|---|
| A | L755S | 9* | SD | 141 |
| B | Y772_A775dup | 6* | SD | 122 |
| C | V777L | 11* | PR | 166 |
| 2 | L313I | 25† | PR | 300 |
| 3 | R456C | 22† | SD | 270 |
| 4 | D769Y | 6† | SD | 180 |
Response of a small cohort of patient with HER2-amplified/mutant tumors from the MSK-IMPACT database performed at the respective hospital as part of routine diagnosis and monitoring.
Response of a small cohort of patient with HER2-amplified/mutant tumors by fluorescence in situ hybridization (FISH) performed at the respective hospital as part of routine diagnosis and monitoring.
Discussion
In this study, we identified somatic mutations of HER2 in tumors from four metastatic HER2-positive patients who had progressed on standard-of-care anti-HER2 therapy. Reviewing 184 HER2-positive patients sequenced at MSKCC, we found a 7.1% prevalence of concomitant amplification and mutation of the HER2 gene (ERBB2); all these patients had metastatic disease that responded poorly to trastuzumab/pertuzumab/paclitaxel combination.
It is known that the presence of HER2 mutations may impair the binding and the activity of lapatinib (5). The catalytic activity, the transforming potential and the sensitivity to kinase inhibitors of HER2 mutations has been traditionally studied expressing the mutant forms of the receptor in ERBB2 non-amplified cells (5, 16). These models, however, do not strictly depend on the exogenous HER2 for proliferation and survival. In order to corroborate our clinical findings and test whether the presence of a HER2 mutation can play a causative role in limiting the sensitivity to anti-HER2 therapy, we strictly utilized ERBB2-amplified models intrinsically sensitive to trastuzumab/lapatinib for our studies. Using a different approach, Xu et al. recently confirmed that the only common somatic mutation gained in BT474 cells chronically exposed to trastuzumab or lapatinib was the HER2 L755S substitution (17), the same encountered in one of our patient at the time of therapy progression. Also in this case, the ectopic expression of this mutation was sufficient to induce resistance to the anti-HER2 agents that were tested. These laboratory findings are in accordance with recent reports showing lack of significant response to trastuzumab-based therapy in HER2-positive metastatic breast cancer patients whose tumors harbor different HER2 mutations (18, 19). Notably, a similar phenomenon has been independently observed by another group that reported an enrichment of HER2 mutations in metastatic lesions from patients undergoing adjuvant trastuzumab (15).
The acquisition of a HER2 activating mutation in the context of HER2 amplification/overexpression may not preclude a response to certain irreversible small molecule HER kinase inhibitors (7, 17). This hypothesis was tested in a clinically relevant animal model where we observed very little control of tumor growth upon trastuzumab or lapatinib treatments, but durable tumor shrinkage following neratinib. More importantly, when we translated these findings into the clinical setting, we observed antitumor activity of neratinib monotherapy (in some cases durable) in breast cancer patients that progressed from anti-HER2 therapies. In the preclinical setting, our studies were limited by the intrinsic difficulty of establishing breast cancer PDXs bearing both ERBB2 genomic alterations. Nevertheless, our in vivo results translated to the clinical setting, showing clinical activity of neratinib in tumors with both amplification and mutations of ERBB2. More patients with similar characteristics will be needed to demonstrate the generalizability of our findings.
In summary, the co-existence of ERBB2 mutations and gene amplification in the metastatic setting may be more frequent than in primary tumors. At this stage, it is still debatable whether this phenomenon is mainly due to the natural evolution of the tumors during the acquisition of metastatic potential or is triggered by anti-HER2 therapy pharmacological pressure. In both cases, these tumors would still depend on the HER2 pathway and may be sensitive to neratinib, which could become a plausible treatment strategy in this setting.
Material and Methods
Patient samples
All patients provided written informed consent for tumor sequencing and review of patient medical records for detailed demographic, pathologic, and previous treatment received. Research protocols for tumor collection and analysis were approved by the ethical committees of Memorial Sloan Kettering Cancer Center, Hospital Universitario Madrid, Candiolo Cancer Institute-FPO and Peter MacCallum Cancer Centre (see text S1). The molecular analysis on archival material was conducted in the context of Institutional Protocol approved by the Internal Review Boards of the Institutions listed in this work. HER2 status was assessed using the HercepTest™ according to the manufacturer’s instructions. HER2 positivity was defined as a 3+ score by immunohistochemistry (IHC) in >10% of invasive tumor cells. Equivocal IHC cases (2 score or 3 score in <10% of invasive tumor cells) were submitted for FISH analysis. A HER2:CEP17 ratio of ≥2.0 was used to define HER2 amplification. The assessment of hormonal receptor status was carried out using IHC with the mAbs against the estrogen receptor (ER) (1/100 dilution; Dako, Glostrup, Denmark) and the progesterone receptor (PgR). The Mib-1 monoclonal antibody (1:200 dilution; Dako) was used to assess Ki-67, which was reported as a percentage of immunoreactive cells among 2,000 tumor cells in randomly selected, high-power fields at the periphery of the tumor. The 184 metastatic breast cancer patients with ERBB2 amplification from MSKCC were identified through prospective clinical genomic profiling between April 2014 and March 2017. Detailed treatment history data were obtained for each patient and included all lines of systemic therapy from time of diagnosis of invasive carcinoma to the study data lock in April 30th, 2017. The exact regimen as well as the dates of start and stop of therapy was recorded for each treatment line.
Capture-based exome sequencing
Unless indicated, multi-gene next generation sequencing was performed using the MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets) assay which involves hybridization of barcoded libraries to custom oligonucleotides (Nimblegen SeqCap) designed to capture all protein-coding exons and select introns of 341 to 468 cancer-associated genes, tumor suppressor genes, and members of pathways deemed actionable by targeted therapies (20, 21). For all the patients, tumor and patient-matched normal DNA samples were extracted from either representative formalin-fixed paraffin embedded (FFPE) tumor biopsy samples or mononuclear cells from peripheral blood. The captured pool was subsequently sequenced on an Illumina HiSeq 2500 as paired-end 100-base pair reads, producing an average of 233-fold coverage per tumor (range = 132x - 453x). Sequence data were analyzed to identify three classes of somatic alterations: single–nucleotide variants, small insertions/deletions (indels), and copy number alterations. Barcoded sequence libraries were prepared using 38 – 250 ng genomic DNA (Kapa Biosystems) and combined in a single equimolar pool. Sequence data were demutliplexed using CASAVA, and reads were aligned to the reference human genome (hg19) using BWA and postprocessed using the Genome Analysis Toolkit (GATK) according to GATK best practices (22, 23). MuTect and GATK were used to call single-nucleotide variants and small indels, respectively (24). Exon-level copy number gains and losses were inferred from the ratio in Tumor:Normal sequence coverage for each target region, following a loess-normalization to adjust for the dependency of coverage on GC content (25).
Cell lines and reagents
BT-474 and Sk-Br-3 HER2-amplified breast cancer cell lines were purchased from ATCC and maintained on DMEM/F12 supplemented with 10% FBS. Wild-type HER2 and HER2 L755S cDNAs were cloned in the pLX302 destination lentiviral vector (ADDGENE #25896). Antibodies against HER2 (#2165), phospho-AKT Ser473 (#4058L), total AKT (#C67E7), phospho-p44/42 MAPK Thr202/Tyr204 (#D13.14.4E), total ERK 1/2 (#9102BC), phospho S6 Ser235/236 (#2211L), total S6 (#2217S) and beta-actin (#4970L) were purchased from Cell Signaling Technology. The phospho-Tyrosine (#05–1050X) antibody was purchased from Millipore. BT-474 and Sk-Br-3 cells were treated with 20 μg/ml of trastuzumab, 500 nM of lapatinib, or the combination of both compounds in the culture media for the times indicated in the figures and/or legends.
Proliferation assays
Clonogenic tumor growth and Cell Titer Glo (CTG) assays were used to assess the in vitro activity of anti-HER2 therapy. For clonogenic tests, 5×104 cells were seeded on 6 cm plates and treated as indicated in the figures and/or legends. Crystal violet staining was performed and experimental replicates were quantified by measuring absorbance at 570 nm on a plate reader. For CTG assays, 5×103 cells/well were seeded in 96-well plates. Twenty-four hours after plating, cells were treated with dimethyl sulfoxide (DMSO) or inhibitors. After 5 days of treatment, CTG assays were conducted according to the manufacturer’s protocol, and dose inhibition curves were produced using the Graphpad Prism software. Comparisons between groups were made using two-tailed Student’s t-test. **P<0.01.
Xenograft models and in vivo studies
PDXs were established from a lung metastasis collected from a breast cancer patient. Preclinical studies were carried out at START (San Antonio, TX) and conducted under International Animal Care and Use Committee (IACUC)-approved protocols. Briefly, tumor fragments were harvested and implanted subcutaneously into the flank of athymic nude mice (Charles River Laboratories). Animals were matched by tumor volume (TV) and randomized to control and treatment groups. TV and animal weight data were collected electronically using a digital calliper and scale; tumor dimensions were converted to volume using the formula TV (mm3) = width2 (mm2) × length (mm) × 0.52. Study endpoint was a mean control TV of approximately 1 cm3; change in TV of each group was compared with the control.
Statistical analysis
We used univariate Cox proportional hazard models to determine the association between HER2 co-mutation/amplification and progression-free survival with disease progression on trastuzumab/pertuzumab/paclitaxel combination therapy or patient death. For patients with multiple lines of therapy, only the first treatment line from that class that was started after the MSK-IMPACT biopsy was included in the analysis. We tested the proportionality assumption of the Cox regression model through time-dependency analysis of selected genetic alterations (cox.zph function of the R package survival). We rejected the null hypotheses with a two-sided α = 0.05. Statistical significance was determined by log-rank test stratified by the treatment.
All the in vivo experiments were run with at least n=8 for each treatment arm. Two-way t-test was performed using GraphPad Prism (GraphPad Software). Error bars represent the SEM.
Supplementary Material
Text S1: Clinical case details
Fig. S1: Prevalence of coincident HER2 amplification and mutation in the MSKCC breast cancer cohort.
Funding:
This work was founded by the NIH grants R03CA187094–01 and P30CA008748, the Breast Cancer Research Foundation, the Geoffrey Beene Cancer Research Center, the Australian New Zealand Breast Cancer Trials Group (ANZBCTG), the FPRC 5 × 1000 Fondi Ministero della Salute grant and from a Puma Biotechnology grant. F.J.C. held a fellowship from the Terri Brodeur Breast Cancer Foundation. EC is a recipient of a MSK Society Scholar Prize.
Footnotes
Competing interests: REC, RPB, FA-C and ASL are employees of Puma Biotechnology. JB has received in the past honoraria from Roche and Novartis. He serves of the board of Varian Medical Systems, Bristol-Myers Squibb and Foghorn and on the scientific advisory boards of Grail, PMV Pharma, Apogen and Northern Biologicals, Tango, and is a founder of Venthera. MS received reserach funds from Puma Biotechnoloy, Daiichi-Sankio and Menarini Ricerche, and is a co-founder of Medendi Medical Travel. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials and further will be made directly available upon request to MS.
References and Notes
- 1.Montemurro F, Scaltriti M, Biomarkers of drugs targeting HER-family signalling in cancer. The Journal of pathology 232, 219–229 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL, Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235, 177–182 (1987). [DOI] [PubMed] [Google Scholar]
- 3.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. , Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 244, 707–712 (1989). [DOI] [PubMed] [Google Scholar]
- 4.Serra V, Vivancos A, Puente XS, Felip E, Silberschmidt D, Caratu G, Parra JL, De Mattos-Arruda L, Grueso J, Hernandez-Losa J, Arribas J, Prudkin L, Nuciforo P, Scaltriti M, Seoane J, Baselga J, Clinical response to a lapatinib-based therapy for a Li-Fraumeni syndrome patient with a novel HER2V659 E mutation. Cancer discovery 3, 1238–1244 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Bose R, Kavuri SM, Searleman AC, Shen W, Shen D, Koboldt DC, Monsey J, Goel N, Aronson AB, Li S, Ma CX, Ding L, Mardis ER, Ellis MJ, Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov 3, 224–237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang SE, Narasanna A, Perez-Torres M, Xiang B, Wu FY, Yang S, Carpenter G, Gazdar AF, Muthuswamy SK, Arteaga CL, HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell 10, 25–38 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Rexer BN, Ghosh R, Narasanna A, Estrada MV, Chakrabarty A, Song Y, Engelman JA, Arteaga CL, Human breast cancer cells harboring a gatekeeper T798M mutation in HER2 overexpress EGFR ligands and are sensitive to dual inhibition of EGFR and HER2. Clin Cancer Res 19, 5390–5401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trowe T, Boukouvala S, Calkins K, Cutler RE Jr., Fong R, Funke R, Gendreau SB, Kim YD, Miller N, Woolfrey JR, Vysotskaia V, Yang JP, Gerritsen ME, Matthews DJ, Lamb P, Heuer TS, EXEL-7647 inhibits mutant forms of ErbB2 associated with lapatinib resistance and neoplastic transformation. Clin Cancer Res 14, 2465–2475 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Shigematsu H, Takahashi T, Nomura M, Majmudar K, Suzuki M, Lee H, Wistuba II, Fong KM, Toyooka S, Shimizu N, Fujisawa T, Minna JD, Gazdar AF, Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res 65, 1642–1646 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Lee JW, Soung YH, Seo SH, Kim SY, Park CH, Wang YP, Park K, Nam SW, Park WS, Kim SH, Lee JY, Yoo NJ, Lee SH, Somatic mutations of ERBB2 kinase domain in gastric, colorectal, and breast carcinomas. Clin Cancer Res 12, 57–61 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Hyman DM, Piha-Paul SA, Won H, Rodon J, Saura C, Shapiro GI, Juric D, Quinn DI, Moreno V, Doger B, Mayer IA, Boni V, Calvo E, Loi S, Lockhart AC, Erinjeri JP, Scaltriti M, Ulaner GA, Patel J, Tang J, Beer H, Selcuklu SD, Hanrahan AJ, Bouvier N, Melcer M, Murali R, Schram AM, Smyth LM, Jhaveri K, Li BT, Drilon A, Harding JJ, Iyer G, Taylor BS, Berger MF, Cutler RE Jr., Xu F, Butturini A, Eli LD, Mann G, Farrell C, Lalani AS, Bryce RP, Arteaga CL, Meric-Bernstam F, Baselga J, Solit DB, HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature 554, 189–194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.N. Cancer Genome Atlas, Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Group M, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S, The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X, De Angelis C, Burke KA, Nardone A, Hu H, Qin L, Veeraraghavan J, Sethunath V, Heiser LM, Wang N, Ng CKY, Chen ES, Renwick A, Wang T, Nanda S, Shea M, Mitchell T, Rajendran M, Waters I, Zabransky DJ, Scott KL, Gutierrez C, Nagi C, Geyer FC, Chamness GC, Park BH, Shaw CA, Hilsenbeck SG, Rimawi MF, Gray JW, Weigelt B, Reis-Filho JS, Osborne CK, Schiff R, HER2 Reactivation through Acquisition of the HER2 L755S Mutation as a Mechanism of Acquired Resistance to HER2-targeted Therapy in HER2(+) Breast Cancer. Clin Cancer Res 23, 5123–5134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuo WJ, Jiang YZ, Wang YJ, Xu XE, Hu X, Liu GY, Wu J, Di GH, Yu KD, Shao ZM, Dual Characteristics of Novel HER2 Kinase Domain Mutations in Response to HER2-Targeted Therapies in Human Breast Cancer. Clin Cancer Res 22, 4859–4869 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Zabransky DJ, Yankaskas CL, Cochran RL, Wong HY, Croessmann S, Chu D, Kavuri SM, Red Brewer M, Rosen DM, Dalton WB, Cimino-Mathews A, Cravero K, Button B, Kyker-Snowman K, Cidado J, Erlanger B, Parsons HA, Manto KM, Bose R, Lauring J, Arteaga CL, Konstantopoulos K, Park BH, HER2 missense mutations have distinct effects on oncogenic signaling and migration. Proc Natl Acad Sci U S A 112, E6205–6214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X, De Angelis C, Burke KA, Nardone A, Hu H, Qin L, Veeraraghavan J, Sethunath V, Heiser LM, Wang N, Ng CKY, Chen ES, Renwick A, Wang T, Nanda S, Shea M, Mitchell T, Rajendran M, Waters I, Zabransky DJ, Scott KL, Gutierrez C, Nagi C, Geyer FC, Chamness GC, Park BH, Shaw CA, Hilsenbeck SG, Rimawi MF, Gray JW, Weigelt B, Reis-Filho JS, Osborne CK, Schiff R, HER2 Reactivation through Acquisition of the HER2 L755S Mutation as a Mechanism of Acquired Resistance to HER2-targeted Therapy in HER2+ Breast Cancer. Clin Cancer Res 23, 5123–5134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boulbes DR, Arold ST, Chauhan GB, Blachno KV, Deng N, Chang WC, Jin Q, Huang TH, Hsu JM, Brady SW, Bartholomeusz C, Ladbury JE, Stone S, Yu D, Hung MC, Esteva FJ, HER family kinase domain mutations promote tumor progression and can predict response to treatment in human breast cancer. Molecular oncology, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boulbes DR, Arold ST, Chauhan GB, Blachno KV, Deng N, Chang WC, Jin Q, Huang TH, Hsu JM, Brady SW, Bartholomeusz C, Ladbury JE, Stone S, Yu D, Hung MC, Esteva FJ, HER family kinase domain mutations promote tumor progression and can predict response to treatment in human breast cancer. Mol Oncol 9, 586–600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Won HH, Scott SN, Brannon AR, Shah RH, Berger MF, Detecting somatic genetic alterations in tumor specimens by exon capture and massively parallel sequencing. J Vis Exp, e50710 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, Hellmann MD, Barron DA, Schram AM, Hameed M, Dogan S, Ross DS, Hechtman JF, DeLair DF, Yao J, Mandelker DL, Cheng DT, Chandramohan R, Mohanty AS, Ptashkin RN, Jayakumaran G, Prasad M, Syed MH, Rema AB, Liu ZY, Nafa K, Borsu L, Sadowska J, Casanova J, Bacares R, Kiecka IJ, Razumova A, Son JB, Stewart L, Baldi T, Mullaney KA, Al-Ahmadie H, Vakiani E, Abeshouse AA, Penson AV, Jonsson P, Camacho N, Chang MT, Won HH, Gross BE, Kundra R, Heins ZJ, Chen HW, Phillips S, Zhang H, Wang J, Ochoa A, Wills J, Eubank M, Thomas SB, Gardos SM, Reales DN, Galle J, Durany R, Cambria R, Abida W, Cercek A, Feldman DR, Gounder MM, Hakimi AA, Harding JJ, Iyer G, Janjigian YY, Jordan EJ, Kelly CM, Lowery MA, Morris LGT, Omuro AM, Raj N, Razavi P, Shoushtari AN, Shukla N, Soumerai TE, Varghese AM, Yaeger R, Coleman J, Bochner B, Riely GJ, Saltz LB, Scher HI, Sabbatini PJ, Robson ME, Klimstra DS, Taylor BS, Baselga J, Schultz N, Hyman DM, Arcila ME, Solit DB, Ladanyi M, Berger MF, Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23, 703–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ, A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G, Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, Ducar M, Van Hummelen P, Macconaill LE, Hahn WC, Meyerson M, Gabriel SB, Garraway LA, High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov 2, 82–93 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Text S1: Clinical case details
Fig. S1: Prevalence of coincident HER2 amplification and mutation in the MSKCC breast cancer cohort.