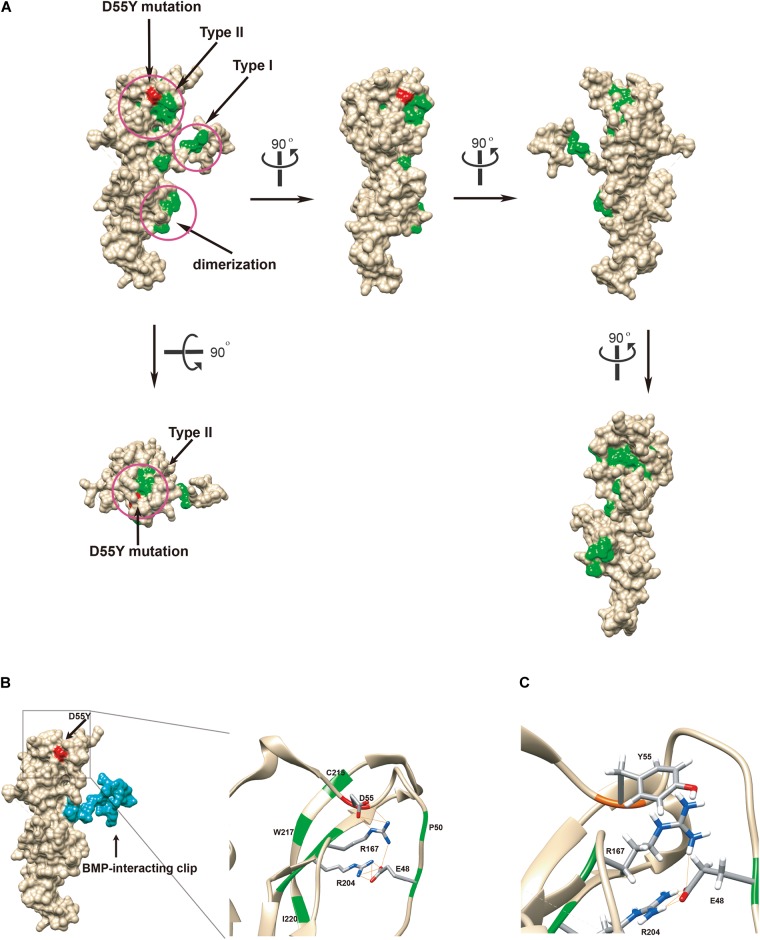
FIGURE 3.
Functional in silico docking analysis of the p.Asp55Tyr mutation in the NOG gene. (A) The disease-associated mutational hotspots on noggin were mapped to show that p.Asp55Tyr is located within the type II receptor binding region. Known mutations are colored green and Asp55 is in red on the Coulombic surface of the protein. Three regions that were previously established are circled. Snapshots of noggin from different point views were taken by rotating the protein 90° at a time. (B) Molecular surface view of noggin is derived from a crystal structure (1M4U). The binding ligand, BMP-14, was removed for clear visualization of noggin. The BMP-interacting clip is colored blue. The location of the mutated residue, Asp55Tyr, is colored red. The type II receptor binding site, where Asp55 resides, is enlarged to show its special arrangement. Among all known mutations, only four residues are hydrogen-bonded (lines colored orange) via side chains to form a network: Glu48, Asp55, Arg167, and Arg204 (the EDRR-tetrad). Asp55 preferentially interacts with Arg167. All residues known to be mutated in diseases are colored green. Detected hydrogen bonds are indicated by lines in orange. (C) In silico generated conformation of the EYRR-tetrad that harbors the p.Asp55Tyr mutation. All three hydrogen bonds are abolished between residue 55 and Arg167. D55Y, Asp55Tyr; C215, Cys215; W217, Trp217; I220, Ile220; D55, Asp55; R167, Arg167; R204, Arg204; E48, Glu48; P50, Pro50; Y55, Tyr55; E48, Glu48.