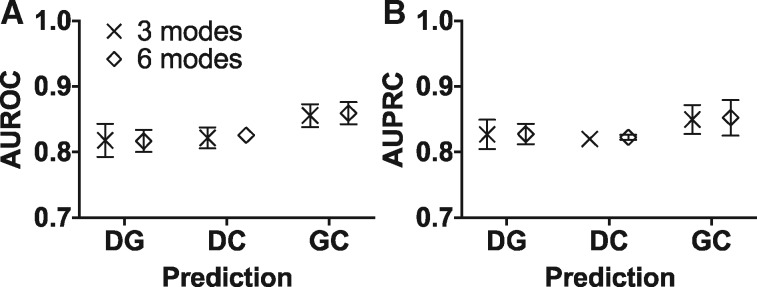
Fig. 2.
Gene-chemical-disease associations can be retrieved by orthogonal types of information in LOMO experiment. The associations of the prediction type in the input network were left out, and then each method used the remaining information to predict them back. (A) AUROC and (B) AUPRC showed similar performance in 3-mode and 6-mode networks. Each dot and its error bar are the mean and the standard deviation of the results from RW, GID and AR. The error bars were not shown if the error bars are shorter than the height of the symbol. The 3 modes, DG+DC+GC network; 6 modes, DG+DC+GC+DD+GG+CC network