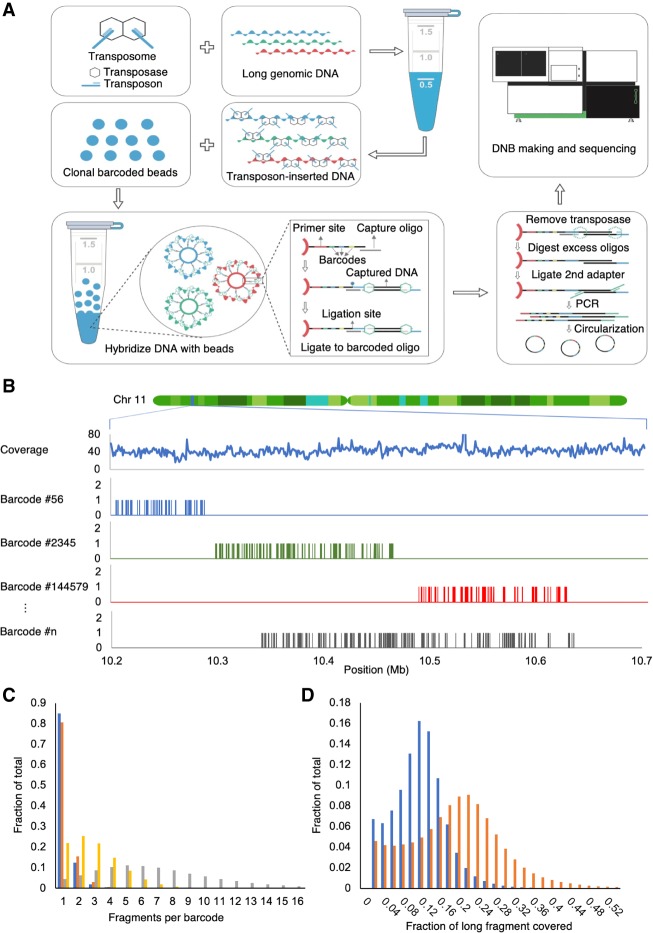
Figure 1.
Overview of stLFR. (A) The first step of stLFR involves inserting a hybridization sequence approximately every 200–1000 bp on long genomic DNA molecules. This is achieved using transposons. The transposon-integrated DNA is then mixed with beads that each contain ∼400,000 copies of an adapter sequence that contains a unique barcode shared by all adapters on the bead, a common PCR primer site, and a common capture sequence that is complementary to the sequence on the integrated transposons. After the genomic DNA is captured to the beads, the transposons are ligated to the barcode adapters. There are a few additional library processing steps and then the cobarcoded subfragments are sequenced on a BGISEQ-500 or equivalent sequencer. (B) Mapping read data by barcode results in clustering of reads within 10- to 350-kb regions of the genome. Total coverage and barcode coverage from four barcodes are shown for the 1-ng stLFR-1 library across a small region on Chromosome 11. Most barcodes are associated with only one read cluster in the genome. (C) The number of original long DNA fragments per barcode are plotted for the 1-ng libraries stLFR-1 (blue) and stLFR-2 (orange) and the 10-ng stLFR libraries stLFR-3 (yellow) and stLFR-4 (gray). More than 80% of the fragments from the 1-ng stLFR libraries are cobarcoded by a single unique barcode. (D) The fraction of nonoverlapping sequence reads (blue) and captured subfragments (orange) covering each original long DNA fragment are plotted for the 1-ng stLFR-1 library.