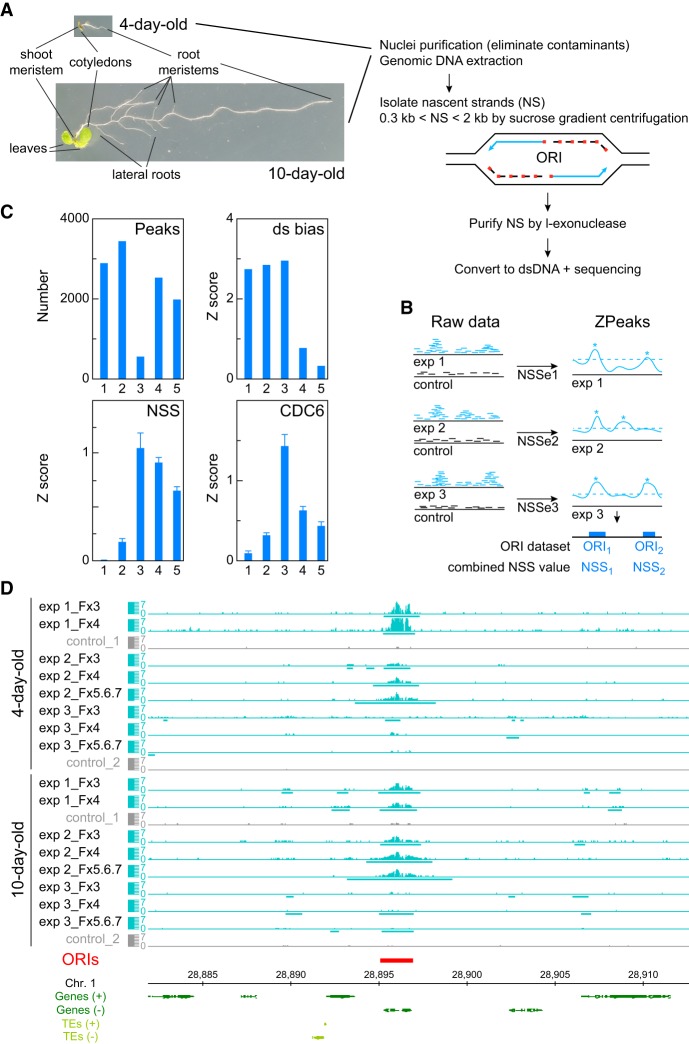
Figure 1.
DNA replication origin (ORI) identification in whole Arabidopsis seedlings and evaluation of reproducibility and quality of sequencing data sets. (A) Summary of basic steps for purification of nascent strands (NS) from seedlings at two developmental stages of Arabidopsis vegetative growth. Seedlings contain cells undergoing cell proliferation and the endocycle in different locations. In 4-d-old seedlings, the shoot and the root apical meristems contain dividing cells, whereas the cotyledons and the transition domain above the root apical meristem contain endocycling cells. The rest of the root is made up of different cell types, some of them dividing and some differentiated. In addition to all these organs and cell types, 10-d-old seedlings contain growing leaf primordia and lateral root primordia, with proliferating cells, as well as a longer root with more differentiated cells. (B) Flowchart summary of the assignment of the nascent strand score (NSS) and the ZPeaks algorithm used to identify ORIs. (C) Quality controls of the NS purification and dsDNA conversion step. The horizontal axis represents the following data sets: (class 1) biased peaks that do not overlap with the ORI set; (class 2) all peaks with dsDNA conversion bias; (class 3) biased peaks overlapping with ORIs; (class 4) all ORIs; and (class 5) ORIs that do not overlap with the biased set. (D) Representative genome browser view of a ∼35-kb region of Chromosome 1 to illustrate ORI identification in various sucrose gradient fractions of the three independent experiments in 4- and 10-d-old seedlings.