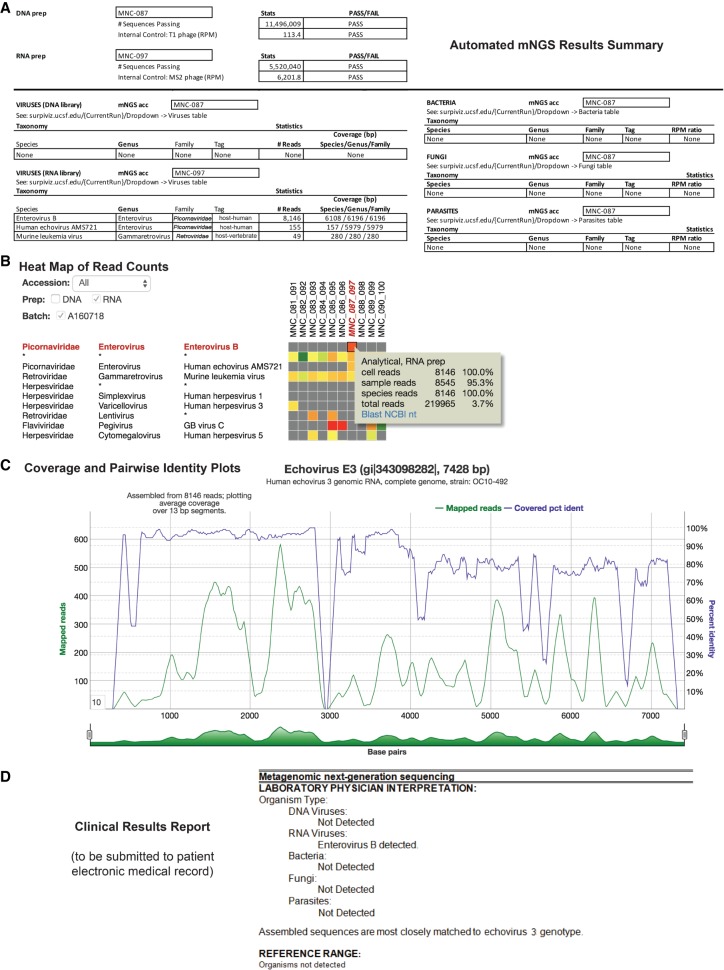
Figure 2.
Identification and reporting of enterovirus infection in a patient with meningoencephalitis using a clinical CSF mNGS assay. SURPI+ provides tools to aid clinical interpretation and graphical visualization (the SURPIviz package), including (A) an automated mNGS results summary, (B) a heat map of aligned reads corresponding to detected pathogens, and (C) a coverage plot (green line) of reads corresponding to a detected pathogen that are mapped to the most closely matched genome or gene in the reference database, along with a corresponding pairwise identity plot (purple line, sliding window = 10 nt). Viral hits corresponding to one CSF patient sample (MNC_087_097, column highlighted in red in B) are taxonomically identified as enterovirus (enterovirus B and echovirus AMS721 species) and murine leukemia virus, a known reagent contaminant (Zheng et al. 2011). After an interpretive review, a laboratory physician prepares a clinical results report (D) that is submitted to the patient electronic medical record (EMR).