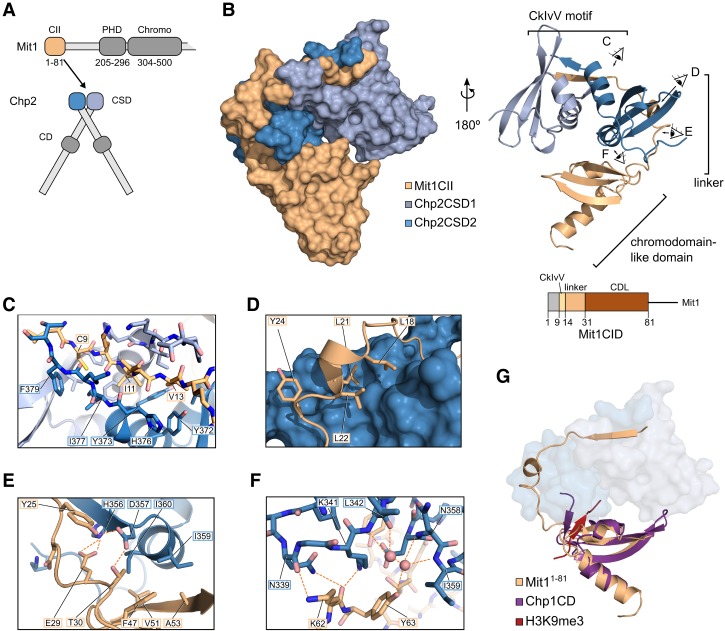
Figure 2.
Crystal structure of the Chp2–Mit1 complex reveals extensive interface. (A) Schematic of the minimal complex between the N-terminal 81 residues of Mit1 and the CSDs of Chp2 based on limited proteolysis (Supplemental Fig. S1A). (B) Surface and cartoon representation of the Chp2–Mit1 crystal structure. Eye symbols with letters indicate viewing angles for corresponding details panels C–F. (C–F) Close-up views of the Chp2–Mit1 crystal structure colored as in (B). (C) Binding of the CkIvV motif to the groove formed by the Chp2CSD dimer. (D) Hydrophobic interactions of the linker region of Mit1 with the surface of Chp2CSD2. (E) Hydrogen bonding network between Mit1 linker domain and Chp2CSD2. (F) Water-mediated Mit1CDL–Chp2CSD2 interaction interface. (G) Superposition of the Mit1CDL domain with the CD of Chp1 bound to a H3K9 trimethyl peptide (PDBID: 3G7L, RMSD = 1.38 Å).