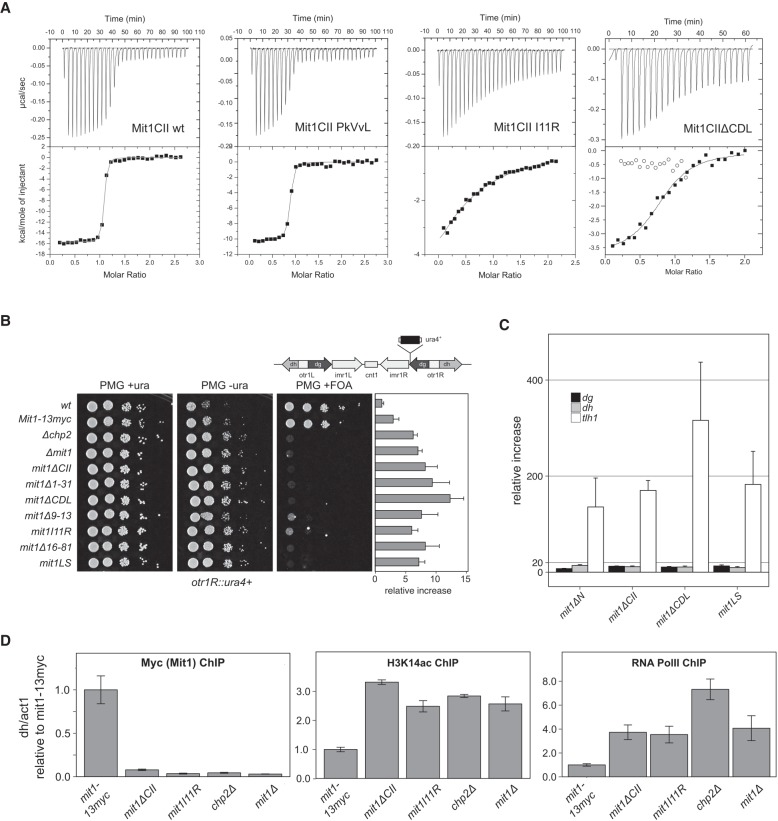
Figure 3.
Extended interaction between Chp2 and Mit1 is required for heterochromatin silencing. (A) ITC heat rates and fits for titration of Chp2CSD with StrepSumo-tagged Mit1CII wild-type and indicated mutant proteins. Open circles represent StrepSumo-tag only control. See Table 2 for fitted parameters. (B) Comparative growth assays for Mit1 mutants in an otr1R::ura4+ background with the corresponding transcript levels of the otr1R::ura4 reporter displayed as horizontal bars measured by quantitative real-time RT-PCR. RT-qPCR levels for wild type, mit1-13myc, Δchp2, and Δmit1 from Figure 1D are shown for comparison (C) Changes in steady-state transcript levels in mutant strains relative to wild-type cells were determined by RT-qPCR for centromeric dg/dh repeats and tlh1 transcripts at telomeres. mit1ΔN from Figure 1D is shown for comparison. (D) Chromatin immunoprecipitation (ChIP) for indicated mutant strains against Mit1-13myc, the H3K14ac histone mark and for RNA polymerase II. act1 was used as internal standard for all measurements and standard errors were calculated from three independent biological experiments.