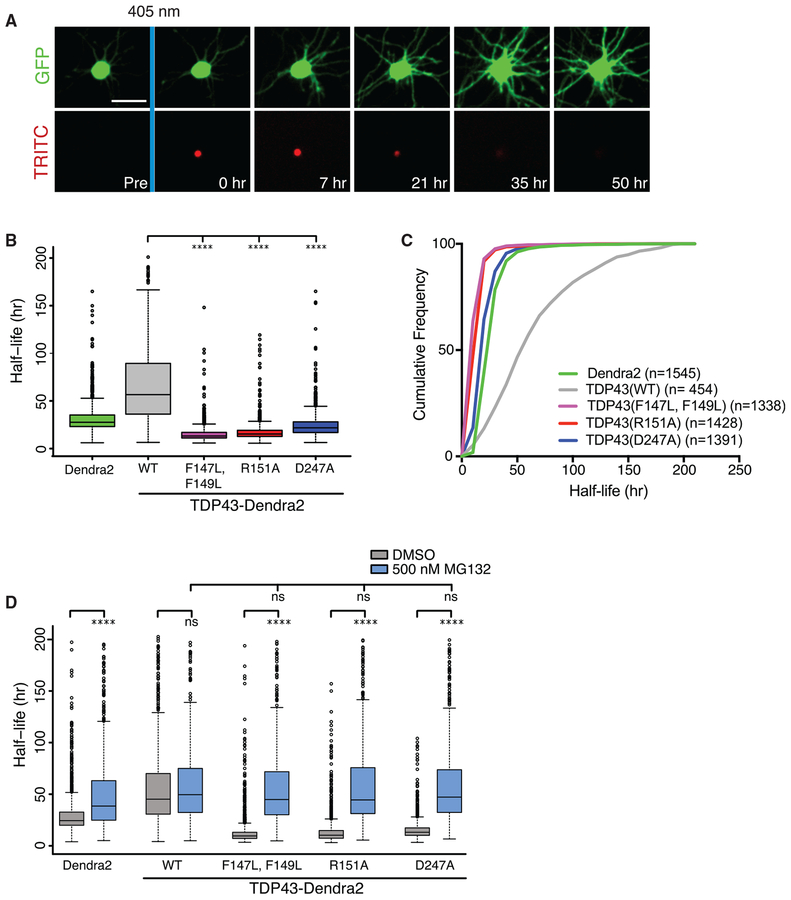
Figure 3. Mutations that Interfere with the RRM1-RRM2 Salt Bridge or RNA Binding Destabilize TDP43.
(A) Optical pulse labeling (OPL) of primary cortical neurons. Neurons transfected with EGFP and TDP43-Dendra2 variants were pulsed with 405 nm of light and imaged by automated microscopy. Scale bar: 50 μm.
(B) The R151A, D247A, and F147L-F149L mutations significantly reduce the half-life of TDP43-Dendra2 ****p < 0.01, 1-way ANOVA with Dunnett’s test.
(C) Cumulative frequency plot of protein half-life data. n, number of neurons.
(D) Six hours before OPL, cells were treated with either DMSO or 500 nM MG132.
****p < 0.01, 1-way ANOVA with Tukey’s test. For (B), data pooled from 4 biological replicates. For (D), data accumulated from 400–2,000 cells per condition, over 3 biological replicates. Plots in (B) and (D) show median (horizontal line), interquartile range (box), and maximum/minimum (vertical lines).