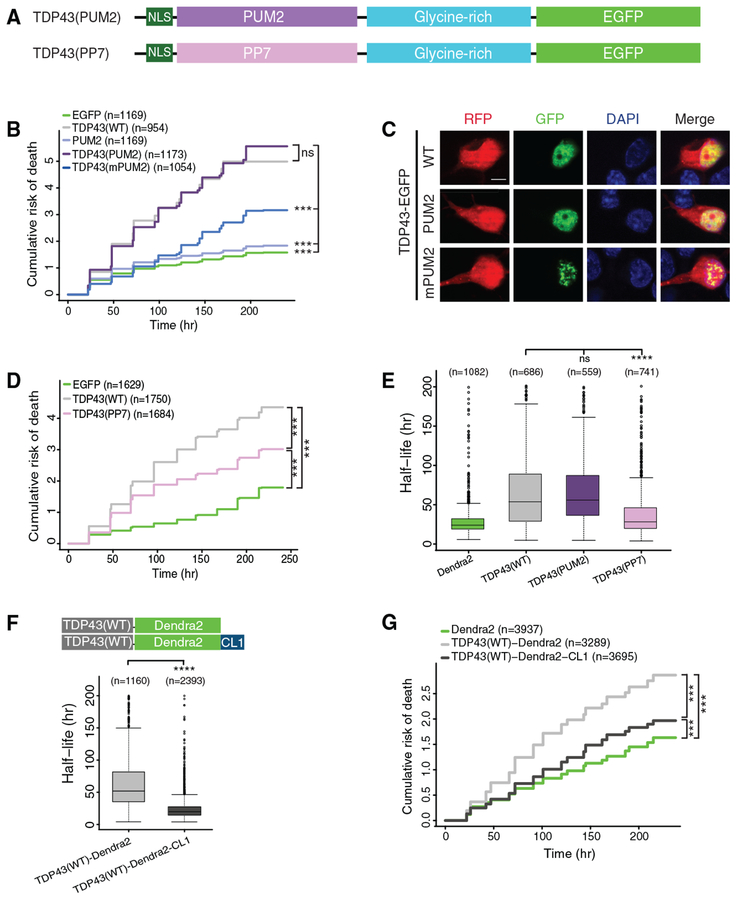
Figure 4. RNA Binding and Protein Stability Are Essential for TDP43-Dependent Neurode-generation.
(A) Schematic of TDP43 hybrid constructs.
(B) TDP43(PUM2)-EGFP expression significantly increased the cumulative risk of death compared to EGFP (HR = 2.54, p < 2 × 10−16). Expression of the PUM2 RNA binding domain (RBD) marginally elevated the risk of death compared to EGFP (HR =1.19, p = 3.78 × 10−5). RNA binding-deficient versions of the PUM2 RBD (mPUM2) significantly reduced the risk of death compared to TDP43(PUM2)-EGFP (HR = 0.45, p < 2 × 10−16).
(C) Images of neurons overexpressing mApple (red fluorescent protein [RFP]) and EGFP-tagged TDP43(WT), TDP43(PUM2), or TDP43(mPUM2). Scale bar: 5 μm.
(D) Expression of TDP43(PP7) reduced the risk of death in comparison to TDP43(WT) (HR = 0.73, p < 2 × 10−16) but remained significantly toxic compared to EGFP (HR = 2.05, p < 2 × 10−16).
(E) Boxplot of TDP43-Dendra2 variant half-life, determined by OPL.
(F) Median TDP43-Dendra2 half-life decreased from 51.9 to 19.8 h with the addition of the destabilizing CL degron.
(G) Both TDP43(WT)-Dendra2 and TDP43(WT)-Dendra2-CL significantly elevated the risk of death compared to Dendra2 (HR = 1.82 and 1.18, p < 2 × 10−16 and p = 1.23 × 10−10, respectively). TDP43(WT)-Dendra2-CL was significantly less toxic compared to TDP43(WT)-Dendra2 (HR = 0.64, p < 2 × 10−16).
Data pooled from 3 independent replicates for (B) and (D) and 4 replicates for (E), (F), and (G). For (B) and (D)–(G), n, number of neurons; ***p < 2 × 10−16; Cox proportional hazards. In (E), ***p < 0.05, 1-way ANOVA with Dunnett’s test. In (F), ****p < 0.0001, unpaired t test. Plots in (E) and (F) show median (horizontal line), interquartile range (box), and maximum/minimum (vertical lines).