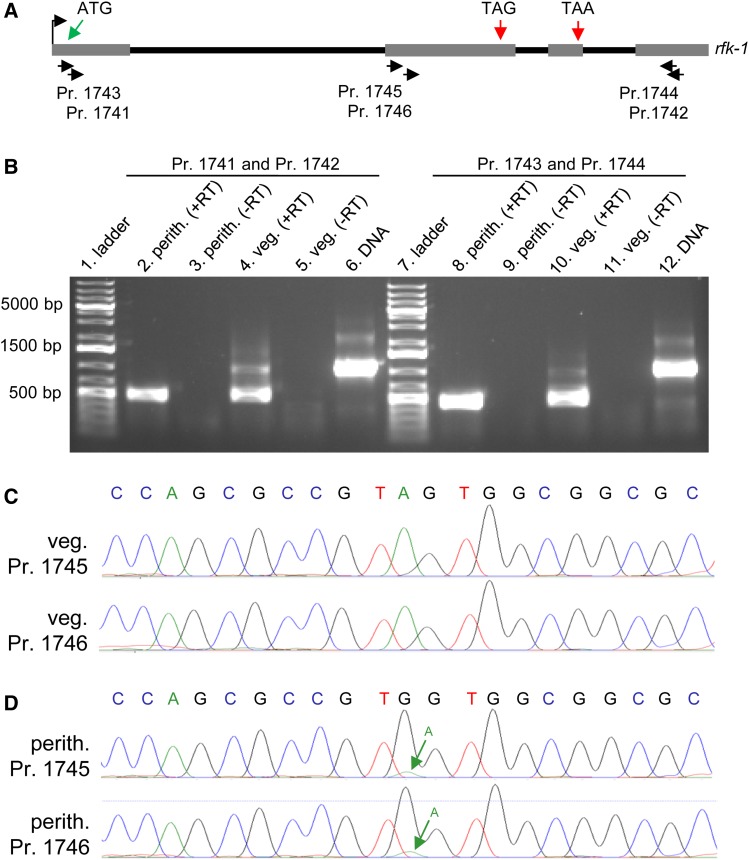
Figure 13.
The rfk-1 mRNA undergoes RNA editing in perithecial tissue. (A) The transcriptional start site (bent arrow), four exons (gray rectangles), and three introns (black lines) of the rfk-1 gene model are depicted in a diagram. The approximate binding sites of six PCR primers (1741 through 1746) within rfk-1 are indicated. (B) RT-PCR analysis was performed on total RNA samples isolated from 6-day-old perithecia of an SkS (F2-26) × Sk-2 (P15-53) cross and from Sk-2 (P15-53) mycelia (36 hr liquid culture). First-strand synthesis was performed with an oligo-dT primer plus reverse transcriptase (+RT). The absence of contaminating DNA was confirmed by leaving reverse transcriptase out of some reactions (−RT). PCR was performed with primers 1741 and 1742 (left) or primers 1743 and 1744 (right), using first-strand cDNA or Sk-2 (P15-53) genomic DNA as template. RT-PCR products were then analyzed by standard agarose gel electrophoresis and visualized with ethidium bromide staining. (C and D) The sequences of the bases surrounding rfk-1 codon 103 [the “TAG” in (A)] within the ∼500-bp cDNA fragments depicted in panel B’s lane 4 and lane 8 were determined directly by Sanger sequencing (without subcloning). (C) Chromatograms from sequencing of the ∼500-bp product from panel B’s lane 4 with primer 1745 or 1746. No editing of codon 103 (TAG) is detected in the chromatograms. (D) Chromatograms from sequencing of the ∼500-bp product from panel B’s lane 8 with primer 1745 or 1746. The green arrow points to a small peak whose presence in the chromatograms suggests that a fraction of the cDNA population contains the original A, instead of the edited G, at this position. Perith., perithecia; Pr., primer; veg., vegetative.