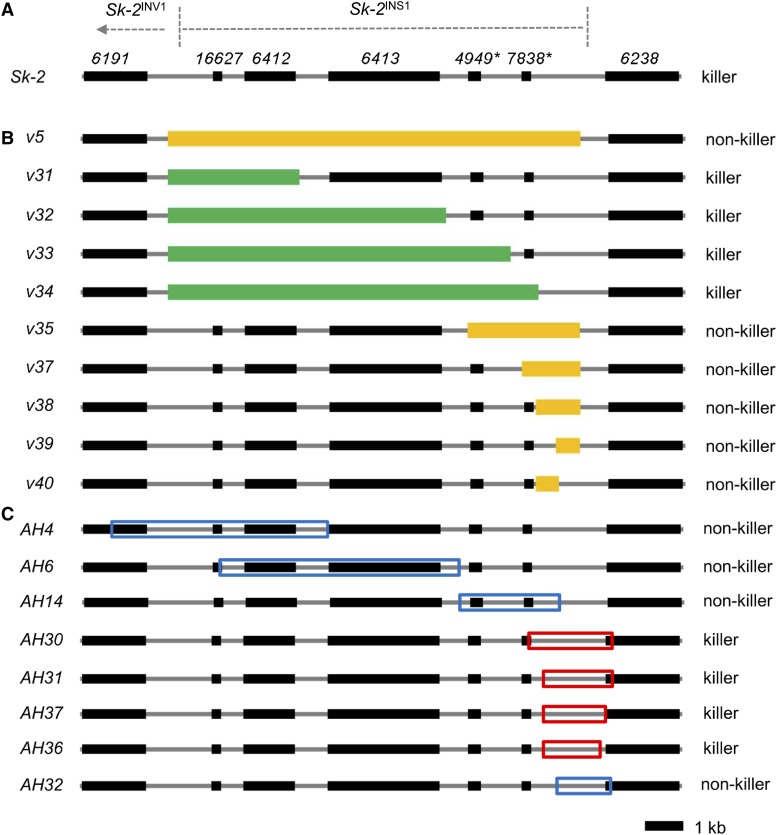
Figure 2.
Deletion and insertion maps. (A) A diagram of Sk-2INS1 and its immediate neighbors. (B) Various intervals of Sk-2INS1 were deleted and replaced with hph. For convenience, each interval was named according to its deletion vector (e.g., interval v5 is named after deletion vector v5). Mango rectangles mark intervals that disrupt spore killing upon deletion. Green rectangles mark intervals that do not disrupt spore killing when deleted. (C) Eight intervals of Sk-2INS1 were transferred to the his-3 locus of an SkS strain. Intervals were labeled according to the name of the plasmid used to clone each interval (e.g., interval AH4 is named after plasmid pAH4). Red and blue boxes mark killer (i.e., abortion-inducing) and nonkiller intervals, respectively.