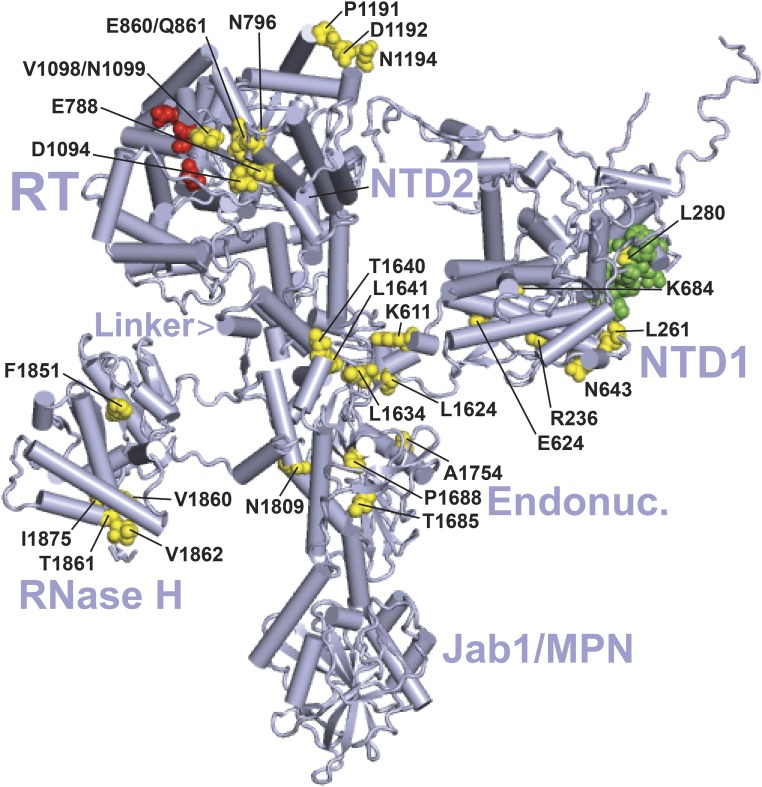
Figure 3.
Location of suppressor substitutions in Prp8 in the yeast B complex. The cryo-EM structure is from Plaschka et al. (2017) (PDB: 5nrl). The major domains of Prp8 are indicated as follows: NTD1 and NTD2, RT, Linker, Endonuclease-like (Endonuc.), RNase H-like (RNase H), and Jab1/MPN (Galej et al. 2013; Nguyen et al. 2016; Bertram et al. 2017). Residues altered by U4-cs1-suppressors identified in Kuhn and Brow (2000) or this study (yellow, Table S6) are labeled if clearly visible. Sites of identified prp28-1 suppressors (Price et al. 2014) and brr2-1 suppressors (Kuhn et al. 2002) are colored green and red, respectively. See Table S6 for substitutions. This and subsequent figures were created using the PyMOL Molecular Graphics System, version 1.8.2.1 (Schrödinger, LLC).