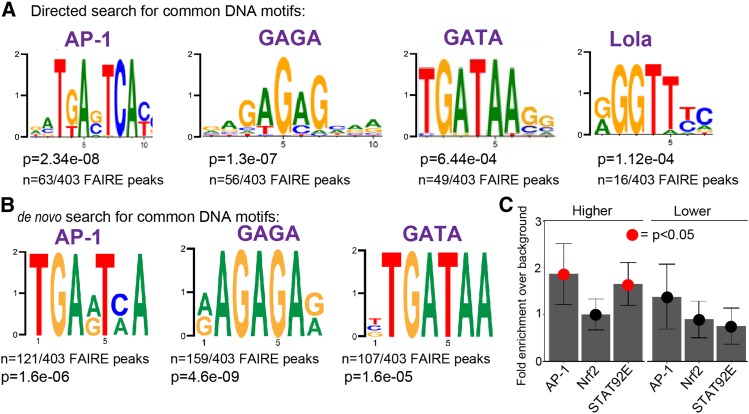
Figure 4.
Enrichment of genes near AP-1, GAGA, GATA, Lola, and STAT92E transcription factor-binding sites. (A) Transcription factor DNA-binding motifs significantly enriched in open chromatin sites near to genes upregulated in both sas-4 and asl. P-values (rank sum test) and fraction of open chromatin peaks containing a designated motif are shown. (B) A de novo motif discovery of open chromatin peaks also uncovered enrichment of AP-1-, GAGA-, and GATA-binding sites. (C) Directed analysis of open chromatin regions near the up- and downregulated genes common to sas-4 and asl for enrichment of binding sites for AP-1, Nrf2, and STAT92E. Data are plotted as the enrichment over genomic background for each motif. Error bars represent 95% C.I.s. Red dots indicate a P-value (Z-test) < 0.05. AP-1 was included as a control for this approach, since we knew from our library-based directed search and the de novo search that AP-1-binding sites should be significantly enriched in our upregulated set of genes. We did not detect significant enrichment for Nrf2 sites using this approach. However, we did detect significant enrichment of the STAT92E-binding motif in our upregulated genes [this motif was not in the library for our directed search, represented in (A)].