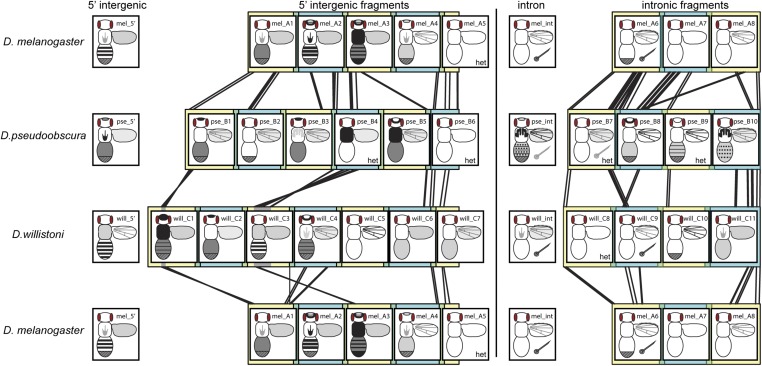
Figure 7.
Synthesizing enhancer activity and sequence similarity for yellow fragments. Overlapping fragments from the 5′ intergenic and intronic regions of D. melanogaster, D. pseudoobscura, and D. willistoni yellow are shown in alternating yellow and blue square blocks, with regions of overlap shown in green. Black lines between fragments from different species show regions of significant sequence similarity detected with the promoterwise program, as described in the Materials and Methods section. Gray regions in the will_C1 and will_C3 fragments indicate sequences that appear to be inverted in D. willistoni relative to the other two species. Enhancer activity observed from each of these fragments, as well as the full 5′ intergenic and intronic sequences from each species, are represented in schematics similar to that shown in Figure 2A. Regions of the body shaded dark gray or black showed higher expression levels than regions of the body shaded light gray. The checkered region at the posterior end of the abdomen indicates sexually dimorphic expression with elevated expression in abdominal segments A5 and A6 of males. The posterior region is fully darkened in the schematic for fragment mel_A3 because it drove elevated expression in abdominal segments A5 and A6 of both males and females. The six fragments assayed only in flies heterozygous for the reporter gene are marked with “het.”