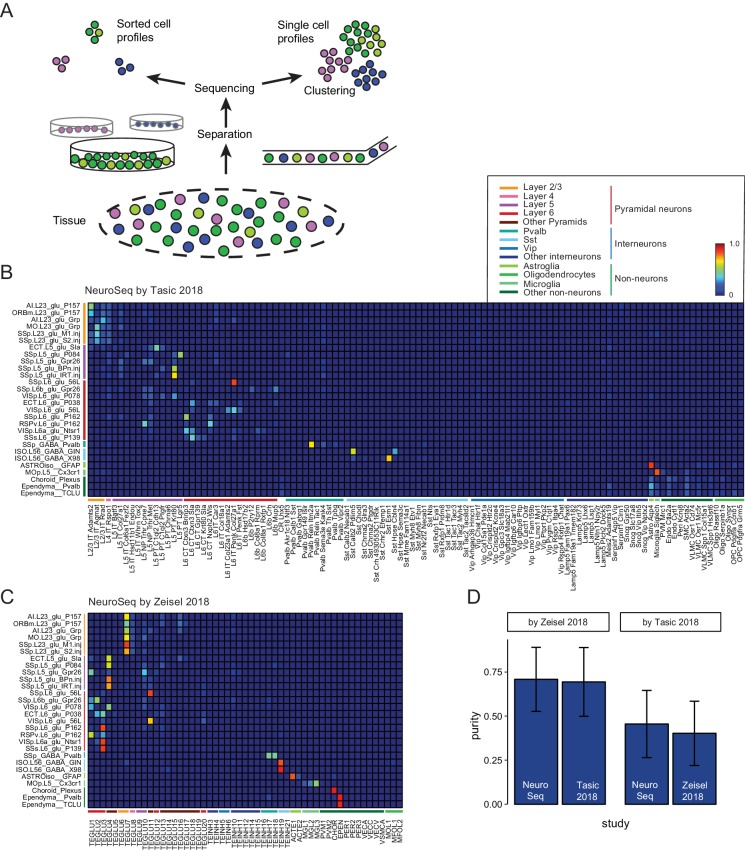
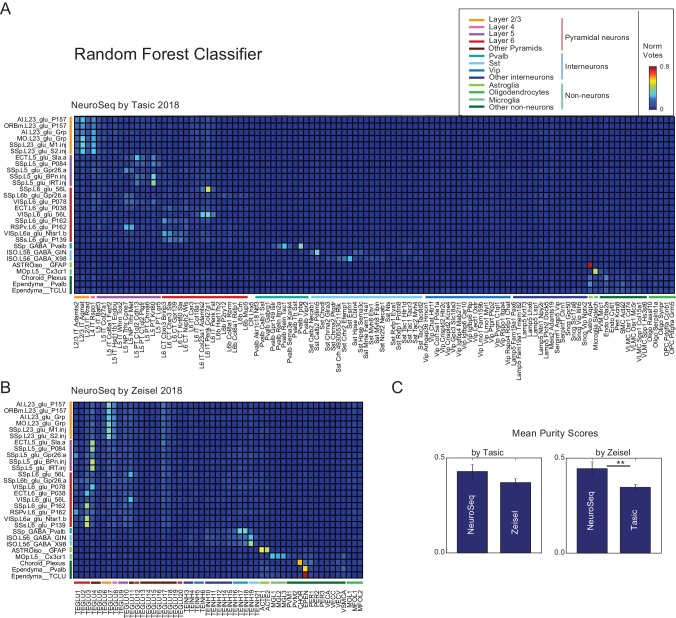
Figure 2. Decomposition by non-negative least squares (NNLS) fitting.
(A) Diagram illustrating potential sources of heterogeneity at the separation phase in profiles from sorted cells (left) or at the clustering phase in profiles from single cells (right). (B,C) NNLS coefficients of NeuroSeq cell populations decomposed by two scRNA-seq datasets: (Tasic et al., 2018; Zeisel et al., 2018). (D) Mean purity scores for NeuroSeq and SC datasets. The purity score for a sample is defined as the ratio of the highest coefficient to the sum of all coefficients. Error bars are Std. Dev.
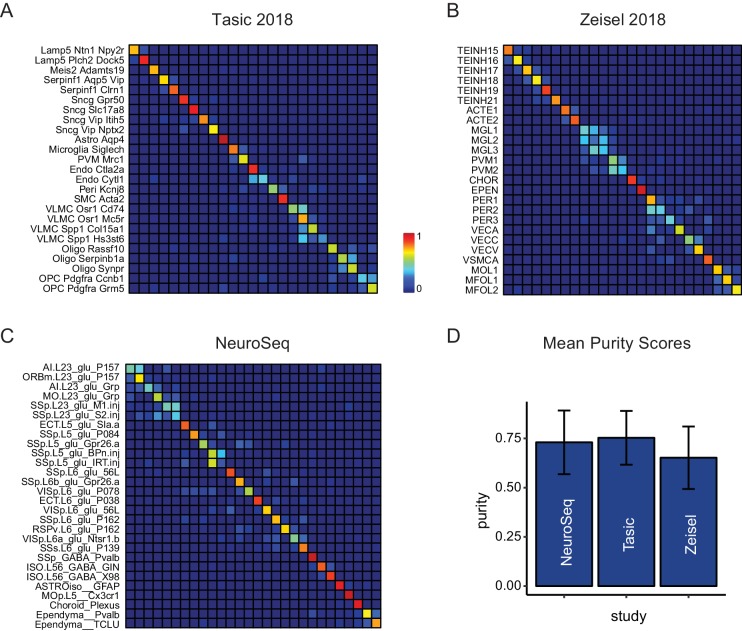
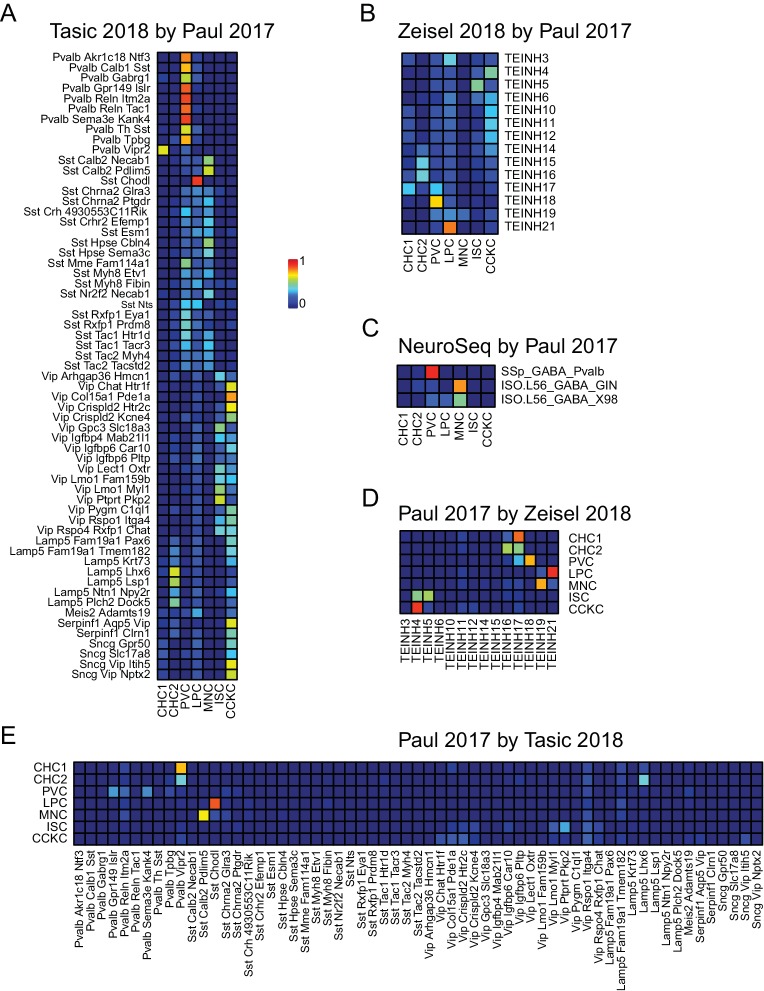
Figure 2—figure supplement 1. Self decompositions by NNLS.
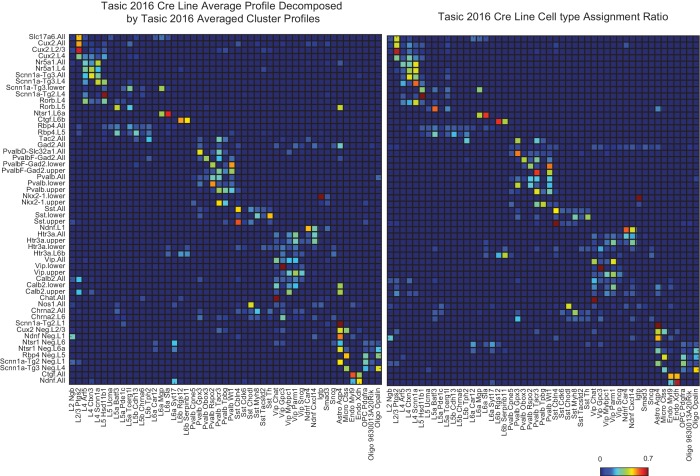
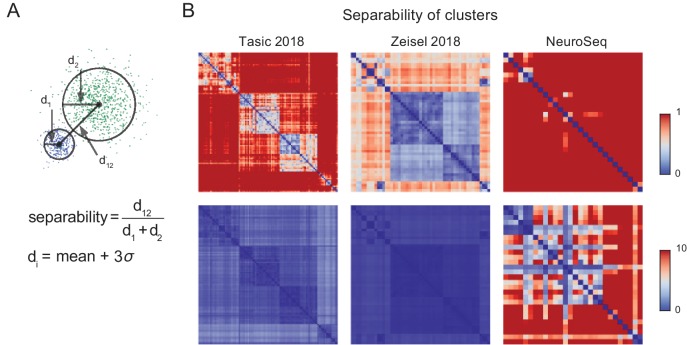
Figure 2—figure supplement 2. A validation of NNLS decomposition.
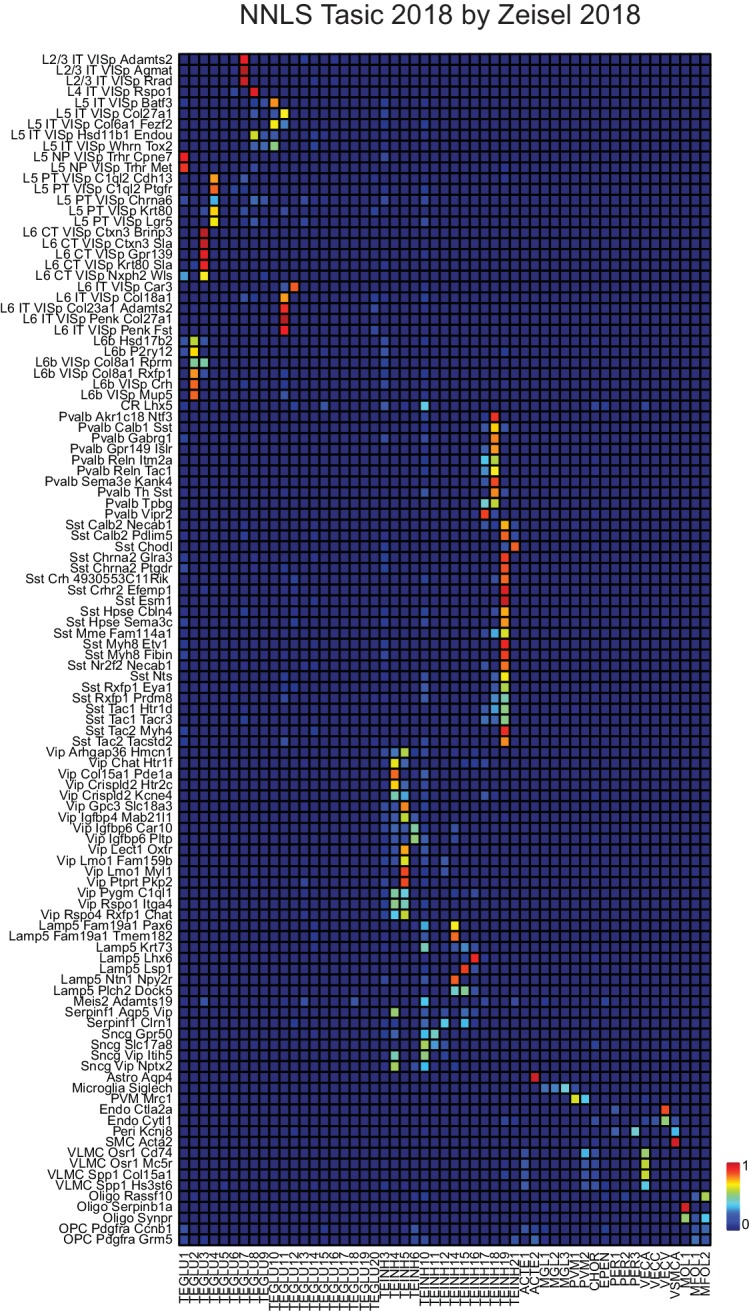
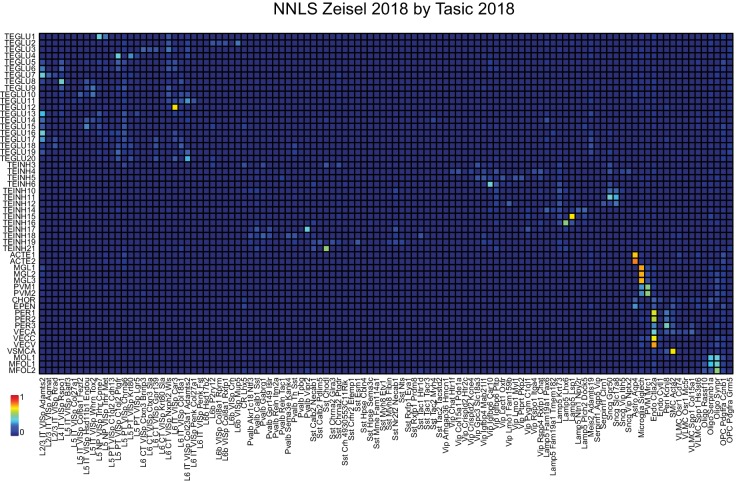
Figure 2—figure supplement 3. NNLS decomposition of SC datasets: Tasic by Zeisel.