Figure 1.
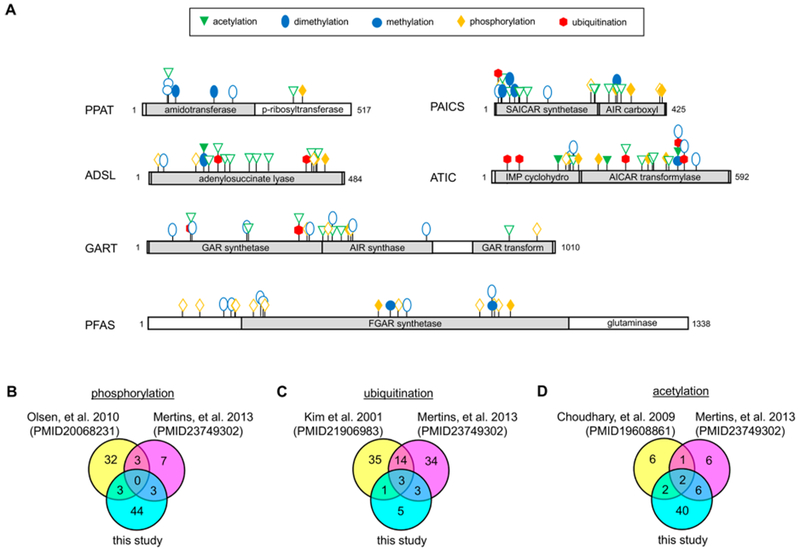
Unambiguously assigned PTMs across the six de novo purine biosynthetic enzymes showing a preference for either purine supplemented or depleted growth conditions. (A) PTMs showing a preference for purine supplemented conditions are denoted by a solid filled symbol corresponding to the type of modification, whereas those observed under purine depleted conditions are represented as outlines of the designated symbol. Unambiguity was determined by a SLIP score of ≥6 for a given modification site. A comparison of our assigned PTMs from this data set to those data sets previously published from global (B) phosphorylation,33,34 (C) ubiquitination,32,33 and (D) acetylation33,35 proteomic studies.
