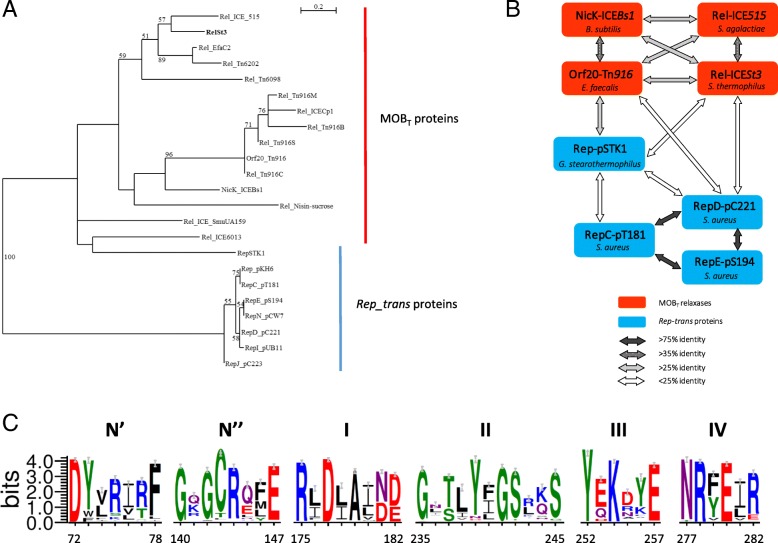
Fig. 1.
Relationships between Rep_trans RCR initiators and MOBT relaxases. a Unrooted maximum likelihood tree of MOBT and Rep_trans proteins. Fifty-four homologous positions were selected manually for phylogenetic analysis by PhyML. Bootstrap values (100 replicates) above 50% are indicated. The scale bar shows the number of substitutions per site. MOBT proteins: RelSt3 encoded by ICESt3 from S. thermophilus [89], Rel_ICE_515 encoded by ICE_515_tRNALys from S. agalactiae [40], Orf20_Tn916 encoded by ICE Tn916 from Enterococcus faecalis [32], NicK_ICEBs1 encoded by ICEBs1 from B. subtilis [33], Rel_Tn6202 encoded by Tn6202 from E. faecalis, conferring vancomycin resistance [41], Rel_ICE_SmuUA159 encoded by ICE_SmuUA159_tRNAleu from S. mutans, Rel_Tn6098 encoded by Tn6098 from Lactococcus lactis, and 4 other Tn916 relaxases from strains of different phyla: Rel_Tn916B from Bifidobacterium longum, Rel_Tn916M from Mycobacterium abscessus, Rel_Tn916S from Streptomyces alboniger, and Rel_Tn916C from Chlamydia trachomatis. Rep_trans proteins: RepSTK1 corresponds to the Rep_trans RCR initiator encoded by the plasmid pSTK1 from Geobacillus stearothermophilus [90], whose structure have been solved recently (PDB ID 4CIJ) [34]. Besides the well-characterized RepC (pT181) and RepD (pC221) proteins, Rep_trans proteins from 5 other members of the pT181 family of staphylococcal plasmids were included: RepE (pS194), RepI (pUB112), RepJ (pC223), RepN (pCW7) and the Rep from pKH6 plasmid [46]. b MOBT relaxases (red boxes) and Rep_trans proteins (blue boxes) are linked according to their global sequence identity (see the bottom panel). c. Weblogos of the six conserved motifs identified for MOBT relaxase domain. 2016 sequences recovered through CDART analysis with RelSt3 sequence as a query were clustered with a sequence identity of 90%, and this WebLogo was obtained using a representative of each of the 261 clusters obtained. The Chemistry color scheme was chosen for representation of amino-acid residues (WebLogo 3 application): green for polar, purple for neutral, blue for basic, red for acidic, and black for hydrophobic residues. Each motif is numbered on the top, and the motif boundaries in the RelSt3 sequence are indicated on the bottom. See Material and Methods for details