Fig. 4.
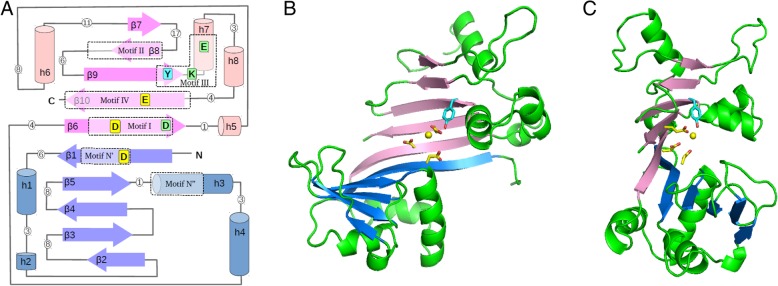
Homology 3-D model of RelSt3 core-domain. a Schematic view of the secondary structure of RelSt3 core domain (PF02486). Beta-strands are represented as arrows and helices as cylinders, with lengths proportional to the numbers of residues. The N- and C-lobes, each including a 5-strand beta-sheet, are coloured in blue and pink, respectively. Loops are represented as grey lines, with white circles indicating their length (number of residues). The five motifs conserved both in MOBT and Rep_trans proteins are boxed in dotted rectangles. In addition, the motif N″ conserved in MOBT proteins identified in this work is also boxed. The residues mutated in this work are indicated and coloured as follows: yellow for residues chelating the divalent cation, blue for the catalytic conserved tyrosine, green for other conserved residues. b. Front view of the homology 3-D model of RelSt3 core-domain. Beta-strands are coloured as in Fig. 4a. Helices and loops are in green. Important residues of the active site are represented as sticks and coloured as in Fig. 4a. c side view of the 3-D RelSt3 model, illustrating the crescent-shaped structure with the active site in the centre. The yellow ball indicates the probable location of the divalent cofactor, according to the model