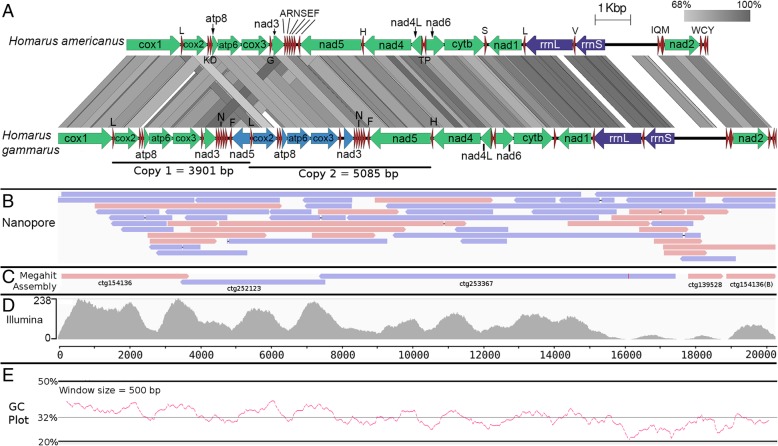
Fig. 3.
Tandem duplication in the H. gammarus mitogenome. a Linear gene organization comparison of the H. gammarus mitogenome with that of H. americanus (tblastx) showing broadly similar gene organization except for the presence of a tandemly duplicated region (cox2-atp8-atp6-cox3-nad3-nad5) in H. gammarus with dissimilar length. The direction of arrow indicates transcription orientation. Green, blue, purple and red arrows correspond to protein-coding genes, putative pseudogenes, rRNAs and tRNAs, respectively. b Minimap2 alignment of nanopore reads to the mitogenome showing broadly even read coverage with some extra long reads spanning the duplicated regions. c Minimap alignment of Megahit Illumina-only assembly indicating the presence of contigs corresponding to both duplicated regions presumably due to the presence of substantial nucleotide divergence. d Bowtie2 mapping of Illumina reads to the mitogenome showing uneven coverage that appears to correlate with the (e) positional (500 bp window size) GC-content of the mitogenome