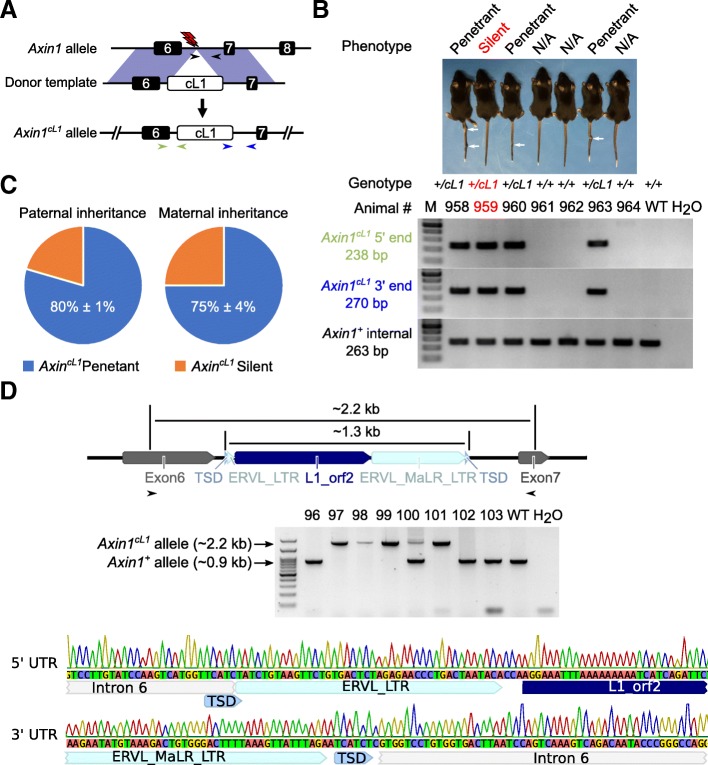
Fig. 1.
Generation of Axin1cL1 mice using CRISPR/Cas9 and phenotypic characterization. a Schematics showing the strategy for generating AxincL1 mice. The red lightning bolt represents the gRNAs used to target the reverse strands of the genomic DNA. The black arrows show the position of internal primers in the Axin1+ allele, whereas the blue and the light green arrows indicate those for amplifying the 5′ and 3’ends of the Axin1cL1 allele, respectively. The expected size of PCR products is indicated in the same color in the lower panels of b. b Image of a representative litter of seven F5 mice derived from a F4 silent AxincL1(Axin1+/cL1) female mouse bred with a WT male, including three penetrant (white arrows pointing to the kinked regions in the tails), one silent AxincL1 mouse and three WT (Axin1+/+) littermates (Upper panel). Genotyping results of these mice are shown in lower panels and the positions of the three sets of primers used are marked in a. c Pie charts showing the distribution of penetrant (with kinky tails) vs. silent (without kinky tails) AxincL1 (Axin1+/cL1) mice in an outbreeding scheme (Axin1+/cL1 × WT) across three generations. Data are represented as means ± SEM (n = 353 for paternal transgenerational inheritance, and n = 425 for maternal transgenerational inheritance). d Confirmation of cL1 insertion into intron 6 of Axin1. Long-range PCR was used to amplify fragments derived from AxincL1 (~ 2.2 kb) and WT (~ 0.9 kb) alleles (upper and middle panels), and the PCR products were sequenced to confirm the successful cL1 insertion in intron 6 of Axin1(lower panels). TSD, target site duplications; ERVL_LTR, LTR of MT2_Mm in ERVL family (5′ extra nucleotide); L1_orf2, orf2 of Lx2 in L1 family; ERVL_MaLR_LTR, LTR of MT_int in ERVL_MaLR family, serving as the 5′ extra nucleotides