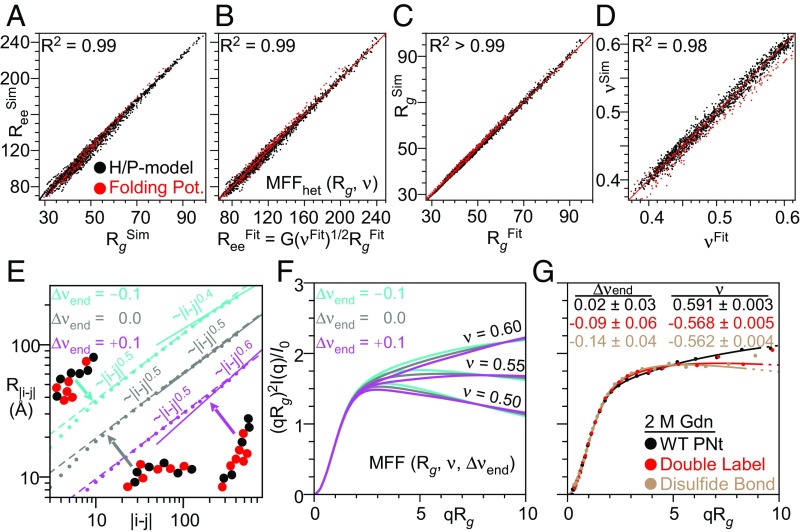
Fig. 4.
Simulations find a strong coupling between Rg and Ree, and SAXS profiles are a robust measure of Rg, ν, and Ree while also informing on the degree of heterogeneity. (A) Coupling between Rg and Ree obtained from the simulated ensembles using the H/P model (black) or our potential used to fold proteins (red). (B–D) Comparison of Rg, ν, and Ree calculated from coordinates of simulated ensembles versus values obtained from fitting with our MFFhet(Rg, ν) to SAXS profiles of the ensembles with randomly added experimental errors. (E) Deviations in νends are observed for heteropolymers with less well-mixed H/P patterns (obtained by a fit to the slope of the dependence of the intrachain distance, R|i − j|, on sequence separation, |i − j| where |i − j| > N/2). (F) Effects of Δνend at different values of ν. (G) Experimental data fit to MFF(Rg, ν, Δνend) demonstrates fluorophore labeling and loop formation via disulfide bonds in PNt induces significant and measurable deviations.