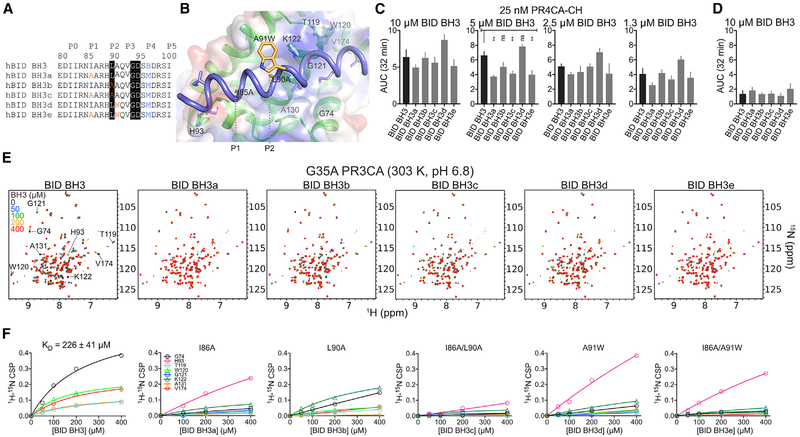
Figure 5. BID BH3 Engages the Canonical Groove of BOK Similar to that of BAK to Promote Liposome Membrane Permeabilization at Specific Dosing Combinations.
(A) Unstapled WT and mutant BID BH3 peptides. Residues highlighted in black are conserved in most BH3-only proteins. Mutations are color coded. B, norleucine.
(B) Cartoon-surface representation of BID BH3 bound to BOK, modeled based on the alignment of BOK NMR structure over the BID BH3-BAK complex (2m5b.pdb). Residues in BID were mutated according to the substitutions in (A). BOK residues that participate in binding have been labeled.
(C) Combined average and SE of AUC at 32 min for WT PR4CA-CH in the presence of unstapled WT and mutant BID BH3 peptides for the normalized LUVP in Figure S5D.
(D) Combined average and SE of AUC at 32 min for WT and mutant BID BH3 peptides alone (Figure S5D).
(E) Overlay of 1H-15N TROSY spectra of 15N-labeled G35A PR3CA titration with WT and mutant BID BH3 at 303 K and pH 6.8.
(F) CSPs plotted against concentration for indicated residues were fitted to a hyperbola to determine the KD for the interaction of G35A PR3CA and the WT BID BH3 peptide. Mutant BID BH3 peptides exhibited weaker binding to BOK compared to the WT.
See also Figure S5.