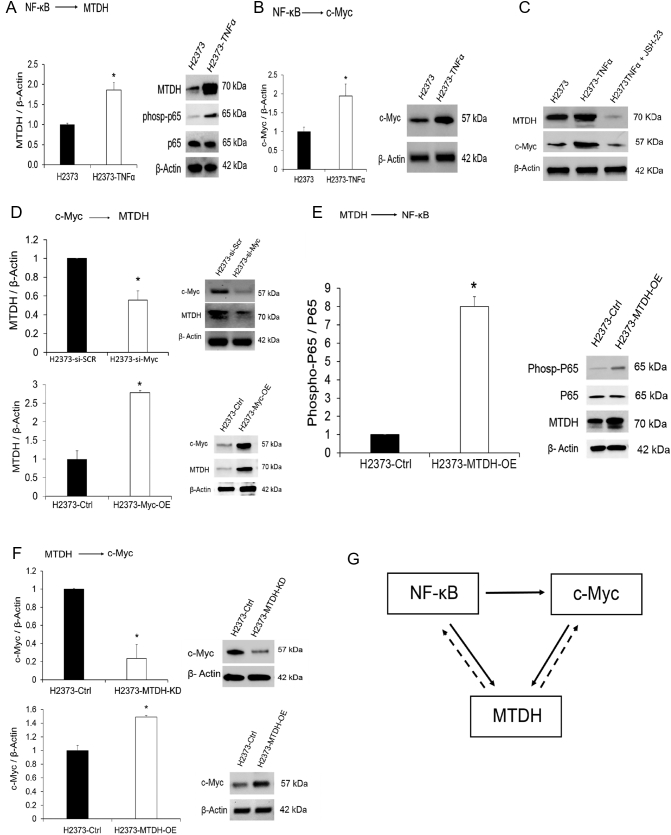
Figure 4.
Metadherin regulation in MPM is influenced by upstream transcription factors.
(A) TNF-α treatment of MPM cells triggers activation of NF-κB signaling as determined by increased phosphorylated p65 protein over a 24-hour duration. MTDH transcript abundance increased (left) as well as protein levels (right). (B) Both c-Myc mRNA (left) and protein (right) expression increased markedly after 24 hours of TNF-α treatment. (C) Treatment of cells with a specific p65 inhibitor (JSH-23) decreased both MTDH and c-Myc protein levels, confirming direct regulation of MTDH and c-Myc by NF-κB. (D) We verified that c-Myc induces MTDH expression. When c-Myc expression was transiently knocked down using siRNA (si-Myc), there was a corresponding decrease observed for both MTDH mRNA (left) and protein (right) levels. Conversely, when c-Myc was overexpressed (Myc-OE), there was a corresponding increase observed for both MTDH mRNA and protein levels. (E) Western blotting confirms that MTDH overexpression results in the activation of NF-κB (p65) in MPM cells, as determined by increased levels of phosphorylated p65 protein. (F) Overexpression and knockdown experiments demonstrate that MTDH positively modulates c-Myc transcript (left) and protein (right) levels in MPM cells. (G) Schematic of a plausible regulatory network controlling MTDH expression in MPM. NF-κB induces feed-forward signaling (solid arrow) that is functionally equivalent to a sensor-coupled switch contributing to maintain activation of MTDH once a threshold of stimulation triggers NF- κB. There are secondary positive feedback loops induced by MTDH itself (dotted arrow). Where applicable, data are presented as mean ± SE. * is P < .05 versus parent cell line and/or negative control specimen. MTDH-OE is stable overexpression of and MTDH-KD is stable gene knockdown of MTDH in MPM cells, respectively.