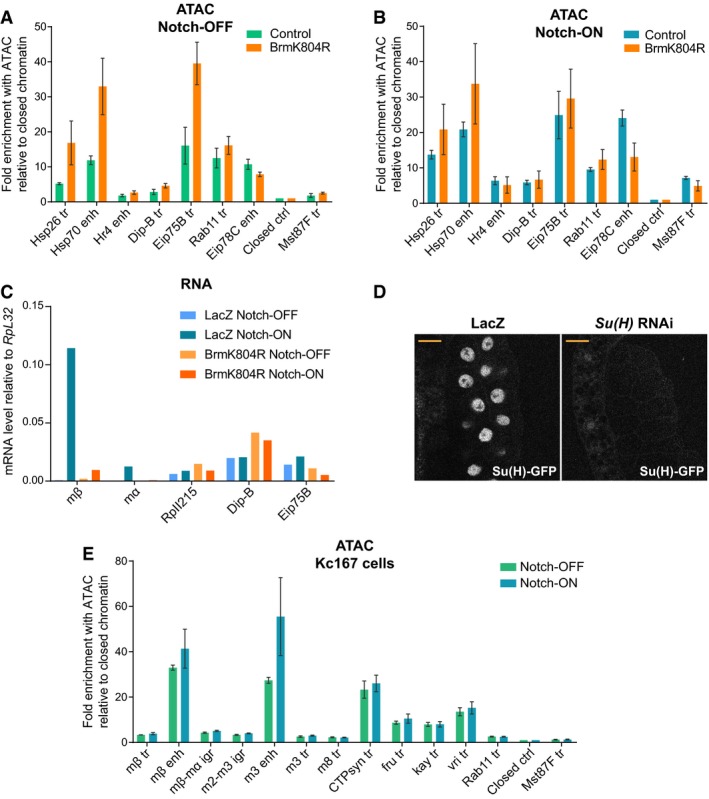
Figure EV3. The effects of BrmK804R on the accessibility and expression of Notch‐inducible genes are not widespread.
-
A, BChromatin accessibility in Notch‐OFF (A) and Notch‐ON (NΔECD expression, B) salivary gland nuclei measured by ATAC‐qPCR; fold enrichment at the indicated regions compared to a “closed ctrl” region. Expression of dominant‐negative Brm, BrmK804R, had little effect or increased the accessibility of some regions. “Hsp26 tr” and “Hsp70 enh” are heat‐shock‐responsive regions, while “Hr4 enh”, “Dip‐B tr”, and “Eip75B” are ecdysone‐responsive. “Rab11 tr”, “Eip78C”, “Closed ctrl”, and “Mst87F tr” are the same as shown in Fig 3B and C. Mean ± SEM; n = 3.
-
CEffect of Notch activation (NΔECD versus LacZ expression) and BrmK804R expression on gene expression in salivary glands measured by reverse transcription–qPCR. Genes shown are Notch‐responsive E(spl)mβ‐HLH (mβ) and E(spl)mα‐HLH (mα), housekeeping gene RpII215, and ecdysone‐responsive Dip‐B and Eip75B. Out of those shown, BrmK804R expression only reduces the Notch‐responsive expression of E(spl)mβ‐HLH and E(spl)mα‐HLH. Mean, n = 2.
-
DEffect of Su(H) RNAi expression on Su(H)‐GFP levels detected with live imaging compared to LacZ expression control. No Su(H)‐GFP was left detectable, and this genotype was used for the ATAC experiment in Fig 3D. Scale bars (yellow) = 50 μm.
-
EAn acute Notch response in Kc167 cells involves increased enhancer accessibility. Chromatin accessibility across the E(spl)‐C in Notch‐ON (EGTA‐treated) and Notch‐OFF (PBS control) Kc167 cells detected by ATAC‐qPCR. Fold enrichment of the indicated regions compared to a “closed ctrl” region; positions of E(spl)‐C primers in the genome are shown in Fig 3A. “CTPsyn tr”, “fru tr”, “kay tr”, and “vri tr” are highly accessible control regions which do not respond to Notch. Mean ± SEM; n = 3.
