Figure 5. Nucleosome turnover at Su(H)‐bound enhancers is increased in Notch‐ON cells and is dependent on the BRM complex.
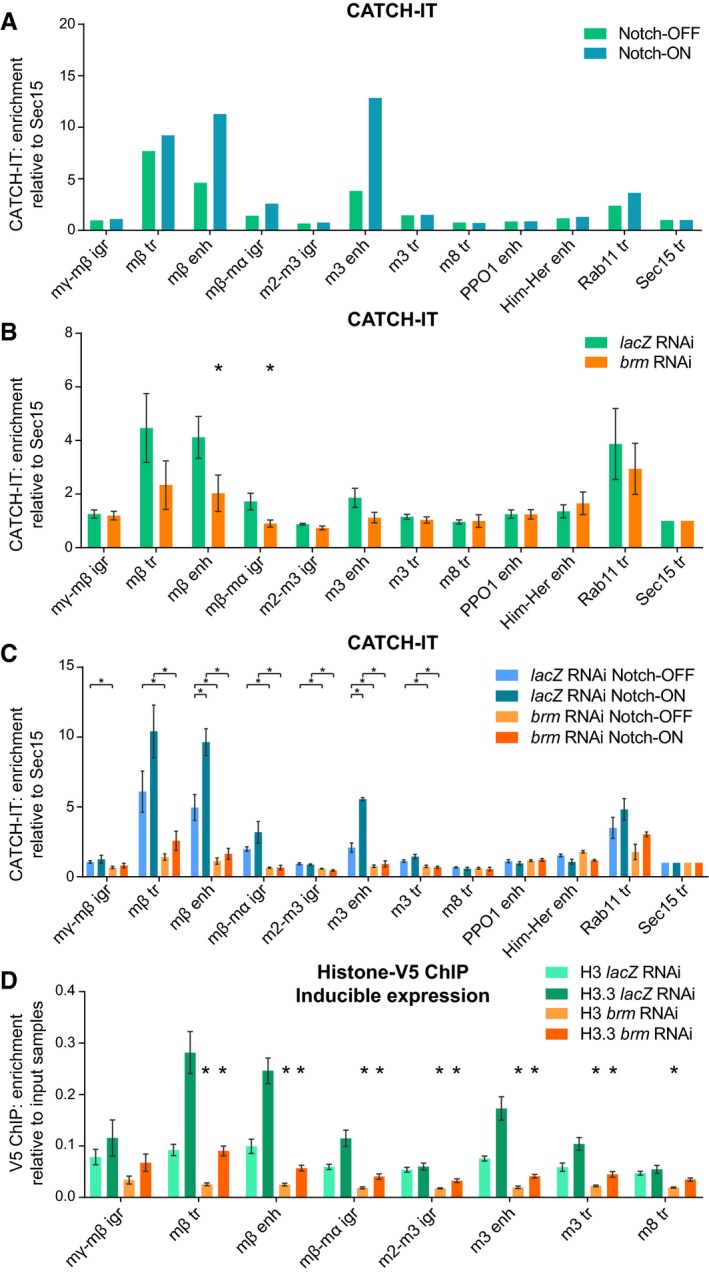
- Nucleosome turnover measured by CATCH‐IT‐qPCR; fold enrichment over input samples compared to Sec15 tr control region. Su(H)‐bound enhancers show increased nucleosome turnover in response to Notch signaling. Notch signaling is activated in Kc167 cells by 6 h of copper induction of pMT‐NICD with copper excluded in the control. Positions of E(spl)‐C primers are shown in Fig 3A; the remaining primers are control non‐Notch‐responsive regions. Mean, n = 2.
- brm RNAi reduces nucleosome turnover at Notch‐responsive regions. CATCH‐IT‐qPCR results as in A after brm or lacZ RNAi as a control. Mean ± SEM; n = 5; *P < 0.05 with one‐tailed Student's t‐test comparing brm and lacZ RNAi.
- Brm is required for Notch‐responsive nucleosome turnover. CATCH‐IT‐qPCR results after brm or lacZ RNAi as in (B) and pMT‐NICD expression as in (A). Mean ± SEM; n = 3; *P < 0.05 with two‐tailed Student's t‐test compared to control (lacZ RNAi Notch‐ON bars are compared to lacZ RNAi Notch‐OFF bars, and brm RNAi bars are compared to their respective lacZ RNAi control bars).
- brm RNAi reduces incorporation of histone H3.3. V5 ChIP–qPCR in Kc167 cells after lacZ or brm RNAi treatment in cells with H3‐V5 or H3.3‐V5 expression induced from the pMT promoter by 3 h of copper treatment, shown as fold enrichment over input samples. Mean ± SEM; n = 3. *P < 0.05 with two‐tailed Welch's t‐test compared to lacZ RNAi control.
