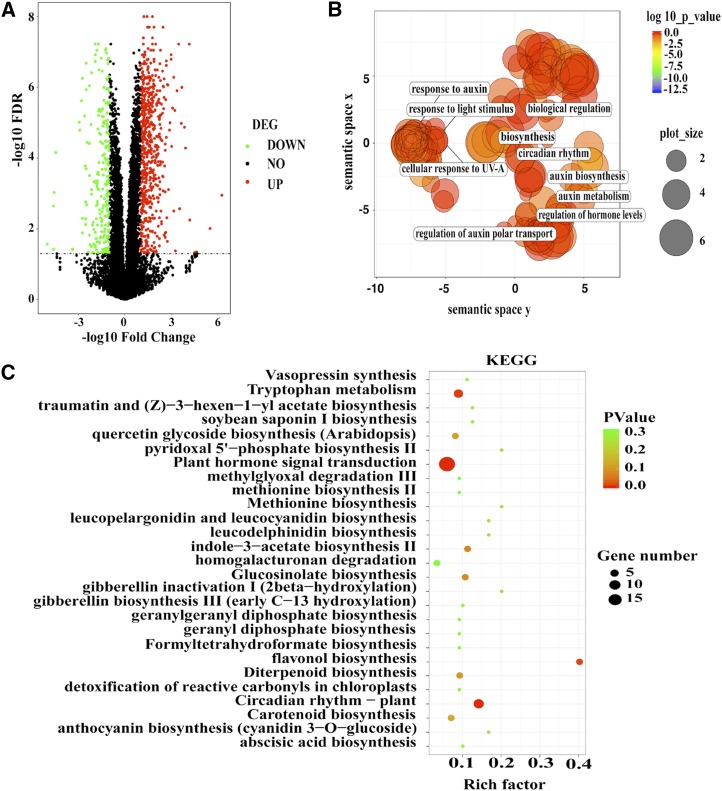
Figure 6.
Globe analysis of target genes regulated by BBX30 and BBX31. A, Volcano plot shows the differentially expressed genes in the bbx30-2 bbx31-2 double mutant. Green spot indicates significant down-regulated genes in the mutant; red spot indicates significantly up-regulated genes; black spot indicates no-change genes. Genes with ratios above 2 or below 0.5. Corrected P value (FDR) ≤ 0.05 was considered as the significant differentially expressed genes (DEG). B, The scatterplot shows the cluster representatives in a two-dimensional space derived by applying multidimensional scaling to a matrix of the enriched GO terms’ semantic similarities (down-regulated genes in the bbx30-2 bbx31-2 double mutant). Bubble color indicates enriched GO term’s P value; size indicates the frequency of the GO term in the GO database (bubbles of more general terms are larger). C, The scatterplot shows the significantly enriched KEGG pathways in the bbx30-2 bbx31-2 double mutant. Bubble color indicates the enriched KEGG pathway’s P value. Significant enriched pathways are marked as red color. Bubble size indicates the gene number in the specific KEGG pathway. Rich factor defines the ratio of input gene number (differentially expressed genes detected in this pathway/background genes of this pathway).